For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRRRAMTKMDEASNPCGGDIEAEMCQLMREQPPAEGVVDRVALQRHRNVALITLSHPQAQNALNLASWRR LKRLLDDLAGESGLRAVVLRGAGDKAFAAGADIKEFPNTRMSAADAAEYNESLAVCLRALTTMPIPVIAA VRGLAVGGGCELATACDVCIATDDARFGIPLGKLGVTTGFTEADTVARLIGPAALKYLLFSGELIGIEEA ARW
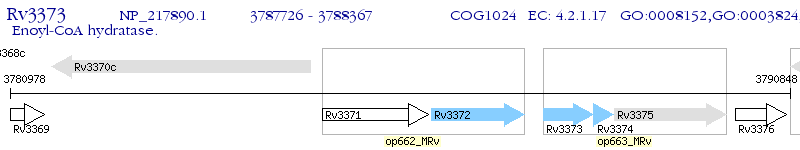
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3373 | echA18 | - | 100% (213) | PROBABLE ENOYL-CoA HYDRATASE ECHA18 (ENOYL HYDRASE) (UNSATURATED ACYL-CoA HYDRATASE) (CROTONASE) |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 7e-26 | 38.51% (174) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0222 | echA1 | 1e-17 | 34.87% (152) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3408 | echA18 | 1e-119 | 100.00% (213) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 3e-25 | 39.08% (174) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 4e-25 | 37.93% (174) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 1e-23 | 36.78% (174) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4396 | echA8 | 2e-26 | 39.08% (174) | enoyl-CoA hydratase EchA8 |
| M. avium 104 | MAV_1195 | - | 2e-26 | 40.23% (174) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 6e-24 | 38.51% (174) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3655 | - | 3e-26 | 38.37% (172) | PUTATIVE short chain enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0210 | echA8 | 1e-26 | 39.08% (174) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4681 | - | 5e-26 | 40.23% (174) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3373|M.tuberculosis_H37Rv MRRRAMTKMDEASNPCGGDIEAEMCQLMREQPPAEGVVDRVALQRHRNVA
Mb3408|M.bovis_AF2122/97 MRRRAMTKMDEASNPCGGDIEAEMCQLMREQPPAEGVVDRVALQRHRNVA
Mflv_2036|M.gilvum_PYR-GCK -----------------------------------MTFETILVDRDDRVA
Mvan_4681|M.vanbaalenii_PYR-1 --------------------------------MGSQNFETIIVERDDRVA
MSMEG_5277|M.smegmatis_MC2_155 ----------------------------------MSDFETILVTRTDRVA
MAB_1185c|M.abscessus_ATCC_199 --------------------------------MTSHNFETILTERIDRVA
MMAR_4396|M.marinum_M -----------------------------------MSYETILVERDQRVG
MUL_0210|M.ulcerans_Agy99 -----------------------------------MSYETILVERDQRVG
MLBr_02402|M.leprae_Br4923 -----------------------------------MKYDTILVDGYQRVG
MAV_1195|M.avium_104 -----------------------------MTDSTDRTFETILVEREERVG
TH_3655|M.thermoresistible__bu ---------------------------------------MVDLELTDGLA
: :.
Rv3373|M.tuberculosis_H37Rv LITLSHPQAQNALNLASWRRLKRLLDDLAGESGLRAVVLRGAGDKAFAAG
Mb3408|M.bovis_AF2122/97 LITLSHPQAQNALNLASWRRLKRLLDDLAGESGLRAVVLRGAGDKAFAAG
Mflv_2036|M.gilvum_PYR-GCK TITLNRPKALNALNTQVMNEVTTAAAELDAEPGVGAIIVTGS-ERAFAAG
Mvan_4681|M.vanbaalenii_PYR-1 TITLNRPKALNALNSQVMDEVTTAAAELDADAGVGAIILTGS-EKAFAAG
MSMEG_5277|M.smegmatis_MC2_155 TIQLNRPKALNALNSQVMTEVTAAAAELDKDPGVGAIIVTGN-EKAFAAG
MAB_1185c|M.abscessus_ATCC_199 VITLNRPKALNALNSQVMNEVTTAAAEFDADHGIGAIIITGS-EKAFAAG
MMAR_4396|M.marinum_M IITLNRPQALNALNSQVMNEVTSAAAELDNDPGIGAIIITGS-AKAFAAG
MUL_0210|M.ulcerans_Agy99 IITLNRPQALNALNSQVMNEVTSAAAELDNDPGIGAIIITGS-AKAFAAG
MLBr_02402|M.leprae_Br4923 IITLNRPQALNALNSQMMNEITNAAKELDIDPDVGAILITGS-PKVFAAG
MAV_1195|M.avium_104 IITLNRPKALNALNTQVMNEVTTAATDFDADPGIGAIIITGS-AKAFAAG
TH_3655|M.thermoresistible__bu VITIDRPQARNAIAPATMEQLEKALDSLAGNPDAQALVIRGAGDRAFVSG
* :.:*:* **: .: .: : . *::: * :.*.:*
Rv3373|M.tuberculosis_H37Rv ADIKEFPNTRMSAADAAEYNESLAVCLRALTTMPIPVIAAVRGLAVGGGC
Mb3408|M.bovis_AF2122/97 ADIKEFPNTRMSAADAAEYNESLAVCLRALTTMPIPVIAAVRGLAVGGGC
Mflv_2036|M.gilvum_PYR-GCK ADIKEMASLSFADVFSSDFFAAWG----KFAATRTPTIAAVAGYALGGGC
Mvan_4681|M.vanbaalenii_PYR-1 ADIKEMAGLSFADVFSSDFFAGWG----KFAATRTPTIAAVAGYALGGGC
MSMEG_5277|M.smegmatis_MC2_155 ADIKEMADLSFAEVFEADFFELWS----KFAATRTPTIAAVAGYALGGGC
MAB_1185c|M.abscessus_ATCC_199 ADIKEMSEQSFSDMFGSDFFSAWG----KLGAVRTPTIAAVSGYALGGGC
MMAR_4396|M.marinum_M ADIKEMADLTFSEAFDADFFAAWG----KLAAVRTPTIAAVAGYALGGGC
MUL_0210|M.ulcerans_Agy99 ADIKEMADLTFSEAFDADFFAAWG----KLAAVRTPTIAAVAGYALGGGC
MLBr_02402|M.leprae_Br4923 ADIKEMASLTFTDAFDADFFSAWG----KLAAVRTPMIAAVAGYALGGGC
MAV_1195|M.avium_104 ADIKEMAELTFADAYGADFFAPWG----KLAALRTPTIAAVAGHALGGGC
TH_3655|M.thermoresistible__bu GDLKELAALRTELEASAMAWRMRS-ICDRLARFPGPVIAALNGHALGGGA
.*:**:. : . : * ***: * *:***.
Rv3373|M.tuberculosis_H37Rv ELATACDVCIATDDARFGIPLGKLGVTTGFTEADTVARLIGPAALKYLLF
Mb3408|M.bovis_AF2122/97 ELATACDVCIATDDARFGIPLGKLGVTTGFTEADTVARLIGPAALKYLLF
Mflv_2036|M.gilvum_PYR-GCK ELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
Mvan_4681|M.vanbaalenii_PYR-1 ELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
MSMEG_5277|M.smegmatis_MC2_155 ELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
MAB_1185c|M.abscessus_ATCC_199 ELAMMCDVIIAAENATFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDMIL
MMAR_4396|M.marinum_M EVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
MUL_0210|M.ulcerans_Agy99 EVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
MLBr_02402|M.leprae_Br4923 ELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
MAV_1195|M.avium_104 ELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLIL
TH_3655|M.thermoresistible__bu EVAVAADIRIAADDIRIGFNQVALAIMPAWGGAERLAELVGRSKALLLAG
*:* .*: **:: :* *.: .. :: ::. :* : :
Rv3373|M.tuberculosis_H37Rv SGELIGIEEAARW-------------------------------------
Mb3408|M.bovis_AF2122/97 SGELIGIEEAARWGLVQKVVAPQDLAAATAKLVGQVCRQSAVTMRAAKVV
Mflv_2036|M.gilvum_PYR-GCK TGRNMDAEEAERAGLVSRVVPADSLLSEAKAVAQTIAGMSLSASRMAKEA
Mvan_4681|M.vanbaalenii_PYR-1 TGRNMGAEEAERAGLVSRVVPADTLLEEARSVAKTIAGMSLSASRMAKEA
MSMEG_5277|M.smegmatis_MC2_155 TGRTIDAAEAERAGLVSRLVPADTLIDEALAVAQTIAGMSLSASRMAKEA
MAB_1185c|M.abscessus_ATCC_199 TGRNMDAAEAERSGLVSRVVATESLLDEAKAVAKTISEMSLSASMMAKEA
MMAR_4396|M.marinum_M TGRTIDAAEAERSGLVSRVVPADDLLTEAKAVATTISQMSRSAARMAKEA
MUL_0210|M.ulcerans_Agy99 TGRTIDAAEAERSGLVSRVVPADDLLTEAKAVATTISQMSRSAARMAKEA
MLBr_02402|M.leprae_Br4923 TGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAKEA
MAV_1195|M.avium_104 TGRTIDAAEAERSGLVSRVVPADDLLTEAKKVATTISQMSRSAARMAKEA
TH_3655|M.thermoresistible__bu TGTVLTAAEAERVGLVDKVVPRASFDTEWRAIARSLATRPAAEIKRGARG
:* : ** *
Rv3373|M.tuberculosis_H37Rv --------------------------------------------------
Mb3408|M.bovis_AF2122/97 ANMHGRAL--TGADTDALIRFGVEAYEGADLREGVAAFSQGRPPKFDD--
Mflv_2036|M.gilvum_PYR-GCK VDRAFETT--LSEGLLYERRLFHSAFATDDQTEGMNAFSEKRSPNFTHR-
Mvan_4681|M.vanbaalenii_PYR-1 VDRAFETT--LTEGLLYERRLFHSAFATDDQTEGMNAFTEKRSPNFTHR-
MSMEG_5277|M.smegmatis_MC2_155 VNRAFETS--LTEGLLYERRLFHSAFATADQKEGMAAFAEKRAANFTHR-
MAB_1185c|M.abscessus_ATCC_199 VNRAFESS--LAEGLLFERRIFHSAFGTADQSEGMAAFVEKRPANFIHR-
MMAR_4396|M.marinum_M VNRTFEAT--LAEGLLYERRLIHSAFATADQSEGMAAFIEKRPPNFTHR-
MUL_0210|M.ulcerans_Agy99 VNRTFEAT--LAEGLLYERRLIHSAFATADQSEGMAAFIEKRPPNFTHR-
MLBr_02402|M.leprae_Br4923 VNRSFEST--LAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR-
MAV_1195|M.avium_104 VNRAFETT--LAEGLLYERRLFHSTFATADQSEGMAAFVEKRAPNFTHR-
TH_3655|M.thermoresistible__bu GRRQPDEPDEVAVGFSPGSRIARSDRRPHNRSAGSAREVPSLRAQLRGQV
Rv3373|M.tuberculosis_H37Rv --------------------------------------------------
Mb3408|M.bovis_AF2122/97 --------------------------------------------------
Mflv_2036|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4681|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5277|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4396|M.marinum_M --------------------------------------------------
MUL_0210|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAV_1195|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu PTGEQPRGRGHPAPDPPPVAGAYQLIVVVTDRVDPVLPVVRAQHQHRVHI
Rv3373|M.tuberculosis_H37Rv --------------------------------------------------
Mb3408|M.bovis_AF2122/97 --------------------------------------------------
Mflv_2036|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4681|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5277|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4396|M.marinum_M --------------------------------------------------
MUL_0210|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAV_1195|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu RARGGPVAVDVDDDVAHAVVGRSHRSVHLRHHLGEGVRIGEGAELVVVDV
Rv3373|M.tuberculosis_H37Rv --------------------------------------------------
Mb3408|M.bovis_AF2122/97 --------------------------------------------------
Mflv_2036|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4681|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5277|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4396|M.marinum_M --------------------------------------------------
MUL_0210|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAV_1195|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu DPVGPAAAEQRPVLLVDTRCVADQYVGDLRPVGVHLSHGNRRGGSRRDHH
Rv3373|M.tuberculosis_H37Rv ----------------------------------------------
Mb3408|M.bovis_AF2122/97 ----------------------------------------------
Mflv_2036|M.gilvum_PYR-GCK ----------------------------------------------
Mvan_4681|M.vanbaalenii_PYR-1 ----------------------------------------------
MSMEG_5277|M.smegmatis_MC2_155 ----------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 ----------------------------------------------
MMAR_4396|M.marinum_M ----------------------------------------------
MUL_0210|M.ulcerans_Agy99 ----------------------------------------------
MLBr_02402|M.leprae_Br4923 ----------------------------------------------
MAV_1195|M.avium_104 ----------------------------------------------
TH_3655|M.thermoresistible__bu RPCNRFSLDENVVSGYCHRRRHRARPPGPGRRGPAGGVNAVVQTPG