For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTARDIMNAGVTCVGEHETLTAAAQYMREHDIGALPICGDDDRLHGMLTDRDIVIKGLAAGLDPNTATAG ELARDSIYYVDANASIQEMLNVMEEHQVRRVPVISEHRLVGIVTEADIARHLPEHAIVQFVKAICSPMAL AS*
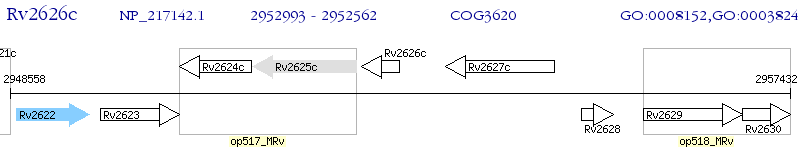
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2626c | - | - | 100% (143) | hypothetical protein Rv2626c |
| M. tuberculosis H37Rv | Rv2406c | - | 1e-05 | 22.32% (112) | hypothetical protein Rv2406c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2659c | - | 6e-79 | 100.00% (143) | hypothetical protein Mb2659c |
| M. gilvum PYR-GCK | Mflv_2670 | - | 2e-06 | 25.89% (112) | signal-transduction protein |
| M. leprae Br4923 | MLBr_00387 | guaB2 | 2e-05 | 23.42% (111) | inosine 5'-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2620 | - | 4e-11 | 35.00% (100) | hypothetical protein MMAR_2620 |
| M. avium 104 | MAV_1776 | - | 9e-06 | 23.21% (112) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4564 | - | 2e-05 | 23.58% (123) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_4428 | - | 2e-13 | 33.90% (118) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3142 | - | 4e-11 | 35.00% (100) | hypothetical protein MUL_3142 |
| M. vanbaalenii PYR-1 | Mvan_1401 | - | 8e-56 | 70.42% (142) | signal-transduction protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu --------------------------------------------------
MLBr_00387|M.leprae_Br4923 MIRGMSNLKESSDFVASSYVRLGGLMDDPAATGGDNPHKVAMLGLTFDDV
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu --------------------------------------------------
MLBr_00387|M.leprae_Br4923 LLLPAASDVVPATADISSQLTKKIRLKVPLVSSAMDTVTEARMAIAMARA
MAV_1776|M.avium_104 ---------------MRIADVLRNKGAAVVTIN------PDATVRELIAG
MSMEG_4564|M.smegmatis_MC2_155 ---------------MRIADVLKNKGTAVVTIS------PQATVTELLAG
Mflv_2670|M.gilvum_PYR-GCK ---------------MRISDVLRSKGAAVATIT------PETSVAGLLTE
Rv2626c|M.tuberculosis_H37Rv ---------------MTTARDIMNAGVTCVG--------EHETLTAAAQY
Mb2659c|M.bovis_AF2122/97 ---------------MTTARDIMNAGVTCVG--------EHETLTAAAQY
Mvan_1401|M.vanbaalenii_PYR-1 ---------------MTTAREIMHTGVTCVG--------EHETLAEAARR
MMAR_2620|M.marinum_M ---------------MTTLPSAGSLSISTVTGDPVARVVADATVADVANA
MUL_3142|M.ulcerans_Agy99 ---------------MTTLPSAGSLSISTVTGDPVARVVADATVADVANA
TH_4428|M.thermoresistible__bu --------------------VVLN---------------PNDPILEAARA
MLBr_00387|M.leprae_Br4923 GGMGVLHRNLPVGEQAGQVETVKRSEAGMVTDPVTCR--PDNTLAQVGAL
. .:
MAV_1776|M.avium_104 LTEQNIGAMVVVGAEG-VLGIVSERDVVRQLHVHGASVLSRPVSKIMTTT
MSMEG_4564|M.smegmatis_MC2_155 LAEMNIGAMVVMGKSG-LEGIVSERDVVRQLHKRGSSLLAQPVSSIMTSV
Mflv_2670|M.gilvum_PYR-GCK LSVRNIGAMVVVSPDG-LQGIVSERDIVRKLHDMGADLLRRPVSEIMTTL
Rv2626c|M.tuberculosis_H37Rv MREHDIGALPICGDDDRLHGMLTDRDIVIKGLAAGLDPNTATAGELARDS
Mb2659c|M.bovis_AF2122/97 MREHDIGALPICGDDDRLHGMLTDRDIVIKGLAAGLDPNTATAGELARDS
Mvan_1401|M.vanbaalenii_PYR-1 MAELGIGALPVCGDDDRLHGMVTDRDIVIKCIAAGHDPAAVTVGELAQGG
MMAR_2620|M.marinum_M MIADNVGAI-VIGDDDRPAALVSERD-VVRVVATGKDPNTVPALDVASTK
MUL_3142|M.ulcerans_Agy99 MIADNVGAI-VIGDDDRPAALVSERD-VVRVVATGKDPNTVPALDVASTK
TH_4428|M.thermoresistible__bu IEENRIGAV-VIQKGGRLVGIVTDRDLAVRALGRGLDPNTTKIAEVMTES
MLBr_00387|M.leprae_Br4923 CARFRISGLPVVDDSGALAGIITNRDMRFEVDQS-----KQVAEVMTKTP
:..: : . .::::** . :
MAV_1776|M.avium_104 LATCTKTDTVDEISVLMTRNRVRHVPVLD-GKKLIGIVSIGDVVKTRMEE
MSMEG_4564|M.smegmatis_MC2_155 VATCTPRDTVDRLNVLMTQNRVRHIPVLD-DGRLAGIVSIGDVVKTRMEE
Mflv_2670|M.gilvum_PYR-GCK VATCTPDDSVDSLSALMTNNRVRHVPVVV-DGRLAGIVSIGDVVKTRMEE
Rv2626c|M.tuberculosis_H37Rv IYYVDANASIQEMLNVMEEHQVRRVPVIS-EHRLVGIVTEADIARHLPEH
Mb2659c|M.bovis_AF2122/97 IYYVDANASIQEMLNVMEEHQVRRVPVIS-EHRLVGIVTEADIARHLPEH
Mvan_1401|M.vanbaalenii_PYR-1 VYHVDADADAQQMLTLMEEHQVRRLPVID-DHRLVGIVSEADVARHLPEY
MMAR_2620|M.marinum_M LVWCDSDATVDQVANQMMEHYIRHVLVEH-DGIPAGIVSARDLLGVYCSD
MUL_3142|M.ulcerans_Agy99 LVWCDSDATVDQVANQMMEHCIRHVLVEH-DGIPAGIVSARDLLGVYCSD
TH_4428|M.thermoresistible__bu PITLTPRDSTDDAIRLMRERNIRRIPLVE-HDRVVGMVTLDDLILDEAAP
MLBr_00387|M.leprae_Br4923 LITAAEGVSADAALGLLRRNKIEKLPVVDGHGRLTGLITVKDFVKTEQHP
: : .. :.:: : *::: *.
MAV_1776|M.avium_104 LEAEQQQLQSYITQG-----------------------------------
MSMEG_4564|M.smegmatis_MC2_155 LETEQQQLQAYITQGR----------------------------------
Mflv_2670|M.gilvum_PYR-GCK LEHEHERLQAYITQG-----------------------------------
Rv2626c|M.tuberculosis_H37Rv AIVQFVKAICSPMALAS---------------------------------
Mb2659c|M.bovis_AF2122/97 AIVQFVKAICSPMALAS---------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 AVAQFVKAICAQQAIA----------------------------------
MMAR_2620|M.marinum_M AVLDSD--------------------------------------------
MUL_3142|M.ulcerans_Agy99 AVLDSD--------------------------------------------
TH_4428|M.thermoresistible__bu LEELAAIVEAQIGEGGPA-------------------DSDRSPARRRRLV
MLBr_00387|M.leprae_Br4923 LATKDNDGRLLVGAAVGVGGDAWVRAMMLVDAGVDVLIVDTAHAHNRLVL
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu RAEATLNRLVKQVQEDAGLEDADQARAALDIVVGALVRRLTPGEAKDFIS
MLBr_00387|M.leprae_Br4923 DMVGKLKVEIGDRVQVIGGNVATRSAAAALVEAGADAVKVGVGPGSTCTT
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu QLPSLLKPHLQTLPPGPDRSVTRESIEGDLVARLGVDQSRATSVLASVAN
MLBr_00387|M.leprae_Br4923 RVVAGVGAPQITAILEAVAACGPAGVPVIADGGLQYSGDIAKALAAGAST
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu TVLDSISPGEADEVRRQLPKELQDVLAAPAAGS-----------------
MLBr_00387|M.leprae_Br4923 TMLGSLLAGTAEAPGELIFVNGKQFKSYRGMGSLGAMQGRGGDKSYSKDR
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Rv2626c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2659c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_2620|M.marinum_M --------------------------------------------------
MUL_3142|M.ulcerans_Agy99 --------------------------------------------------
TH_4428|M.thermoresistible__bu --------------------------------------------------
MLBr_00387|M.leprae_Br4923 YFADDALSEDKLVPEGIEGRVPFRGPLSSVIHQLVGGLRAAMGYTGSPTI
MAV_1776|M.avium_104 ------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 ------------------------------------
Mflv_2670|M.gilvum_PYR-GCK ------------------------------------
Rv2626c|M.tuberculosis_H37Rv ------------------------------------
Mb2659c|M.bovis_AF2122/97 ------------------------------------
Mvan_1401|M.vanbaalenii_PYR-1 ------------------------------------
MMAR_2620|M.marinum_M ------------------------------------
MUL_3142|M.ulcerans_Agy99 ------------------------------------
TH_4428|M.thermoresistible__bu ------------------------------------
MLBr_00387|M.leprae_Br4923 EVLQQAQFVRITPAGLKESHPHDVAMTVEAPNYYPR