For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIADVLKNKGTAVVTISPQATVTELLAGLAEMNIGAMVVMGKSGLEGIVSERDVVRQLHKRGSSLLAQPV SSIMTSVVATCTPRDTVDRLNVLMTQNRVRHIPVLDDGRLAGIVSIGDVVKTRMEELETEQQQLQAYITQ GR*
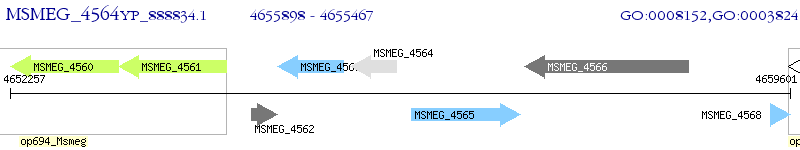
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4564 | - | - | 100% (143) | CBS domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1602 | guaB | 9e-08 | 28.46% (123) | inositol-5-monophosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2429c | - | 4e-55 | 73.94% (142) | hypothetical protein Mb2429c |
| M. gilvum PYR-GCK | Mflv_2670 | - | 9e-51 | 68.31% (142) | signal-transduction protein |
| M. tuberculosis H37Rv | Rv2406c | - | 4e-55 | 73.94% (142) | hypothetical protein Rv2406c |
| M. leprae Br4923 | MLBr_00387 | guaB2 | 3e-08 | 29.27% (123) | inosine 5'-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3721c | - | 5e-09 | 30.89% (123) | inosine 5'-monophosphate dehydrogenase |
| M. marinum M | MMAR_3726 | - | 2e-55 | 75.35% (142) | transcriptional regulator |
| M. avium 104 | MAV_1776 | - | 7e-56 | 74.65% (142) | CBS domain-containing protein |
| M. thermoresistible (build 8) | TH_1455 | - | 1e-43 | 65.91% (132) | CBS domain protein |
| M. ulcerans Agy99 | MUL_3669 | - | 1e-54 | 73.94% (142) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3907 | - | 4e-51 | 70.42% (142) | signal-transduction protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 MIRGMSNLKESSDFVASSYVRLGGLMDDPAATGGDNPHKVAMLGLTFDDV
MAB_3721c|M.abscessus_ATCC_199 ----MT------------------IAAGSVHTGGDDPHKVAMLGLTFDDV
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 LLLPAASDVVPATADISSQLTKKIRLKVPLVSSAMDTVTEARMAIAMARA
MAB_3721c|M.abscessus_ATCC_199 LLLPAASDVLPAGADTSSQLTKRIRLNVPLVSSAMDTVTEARMAIAMARA
Mflv_2670|M.gilvum_PYR-GCK ----------------MRISDVLRSKG---AAVATITPETSVAGLLTELS
Mvan_3907|M.vanbaalenii_PYR-1 ----------------MRISDVLRNKG---ATVATITPETSVAGLLTELS
TH_1455|M.thermoresistible__bu --------------------------------VATVTETTTITGLLAEMA
Mb2429c|M.bovis_AF2122/97 ----------------MRIADVLRNKG---AAVVTINPDATVGELLAGLA
Rv2406c|M.tuberculosis_H37Rv ----------------MRIADVLRNKG---AAVVTINPDATVGELLAGLA
MMAR_3726|M.marinum_M ----------------MRIADVLRNKG---AAVTTINPDATVRELLAGLA
MUL_3669|M.ulcerans_Agy99 ----------------MRIADVLRNKG---AVVTTINPDATVRELFAGLA
MAV_1776|M.avium_104 ----------------MRIADVLRNKG---AAVVTINPDATVRELIAGLT
MSMEG_4564|M.smegmatis_MC2_155 ----------------MRIADVLKNKG---TAVVTISPQATVTELLAGLA
MLBr_00387|M.leprae_Br4923 GGMGVLHRNLPVGEQAGQVETVKRSEAGMVTDPVTCRPDNTLAQVGALCA
MAB_3721c|M.abscessus_ATCC_199 GGMGVLHRNLSVEDQAGQVETVKRSEAGMVTNPVTCSPANTLAEVDALCA
.* :: : : :
Mflv_2670|M.gilvum_PYR-GCK VRNIGAMVVVSPDG---LQGIVSERDIVRKLHDMGADLLRRPVSEIMT-T
Mvan_3907|M.vanbaalenii_PYR-1 VHNIGAMVVVSPDG---LLGIVSERDVVRKLHDMGADLLRRPVSEIMS-T
TH_1455|M.thermoresistible__bu THNVGAMVVVRPDGSGGIAGMVSERDVVRKLHQRGANLLDQPVSEIMT-S
Mb2429c|M.bovis_AF2122/97 EQNIGAMVVVGAEG---VVGIVSERDVVRQLHTYGASVLSRPVAKIMS-T
Rv2406c|M.tuberculosis_H37Rv EQNIGAMVVVGAEG---VVGIVSERDVVRQLHTYGASVLSRPVAKIMS-T
MMAR_3726|M.marinum_M EQNIGAMVVVGVEG---VVGIVSERDVVRQLHTHGASVLTRPVSKIMT-S
MUL_3669|M.ulcerans_Agy99 EQNIGAMVVVGVEG---VVGIVSERDVVRQLHTHGASVLTRPVSKIMT-S
MAV_1776|M.avium_104 EQNIGAMVVVGAEG---VLGIVSERDVVRQLHVHGASVLSRPVSKIMT-T
MSMEG_4564|M.smegmatis_MC2_155 EMNIGAMVVMGKSG---LEGIVSERDVVRQLHKRGSSLLAQPVSSIMT-S
MLBr_00387|M.leprae_Br4923 RFRISGLPVVDDSG--ALAGIITNRDMRFEVD------QSKQVAEVMTKT
MAB_3721c|M.abscessus_ATCC_199 RFRISGLPVVDAQG--ALVGIITNRDMRFEAD------LSKPVAEVMTKA
.:..: *: .* : *::::**: : . : *:.:*: :
Mflv_2670|M.gilvum_PYR-GCK LVATCTPDDSVDSLSALMTNNRVRHVPVVVD-GRLAGIVSIGDVVKTRME
Mvan_3907|M.vanbaalenii_PYR-1 LVATCTPDDTVDSLSALMTNNRVRHVPVVVD-GRLAGIVSIGDVVKTRME
TH_1455|M.thermoresistible__bu LVVTCSPDDAVDDLAAMMTDNRVRHVPVLVD-GRLAGIVSIGDVVKTRLE
Mb2429c|M.bovis_AF2122/97 TVATCTKSDTVDKISVLMTENRVRHVPVLDG-KKLIGIVSIGDVVKSRMG
Rv2406c|M.tuberculosis_H37Rv TVATCTKSDTVDKISVLMTENRVRHVPVLDG-KKLIGIVSIGDVVKSRMG
MMAR_3726|M.marinum_M AVATCTKSDTVDSISVLMTQNRVRHVPVLDG-QKLIGIVSIGDVVKSRMG
MUL_3669|M.ulcerans_Agy99 AVATCTKSDTVDSISVLMTQNRVRHVPVLDG-QKLIGIVSIGDVVKSRMG
MAV_1776|M.avium_104 TLATCTKTDTVDEISVLMTRNRVRHVPVLDG-KKLIGIVSIGDVVKTRME
MSMEG_4564|M.smegmatis_MC2_155 VVATCTPRDTVDRLNVLMTQNRVRHIPVLDD-GRLAGIVSIGDVVKTRME
MLBr_00387|M.leprae_Br4923 PLITAAEGVSADAALGLLRRNKIEKLPVVDGHGRLTGLITVKDFVKTEQH
MAB_3721c|M.abscessus_ATCC_199 PLITAREGVTADAALGLLRRNKIEKLPIVDGEGRLTGLITVKDFAKTEQH
: *. :.* :: *::.::*:: . :* *:::: *..*:.
Mflv_2670|M.gilvum_PYR-GCK ELEHEHERLQAYITQG----------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 ELEREQQHLQAYITQG----------------------------------
TH_1455|M.thermoresistible__bu ELEAEQEQLHAYITGG----------------------------------
Mb2429c|M.bovis_AF2122/97 ELEAEQQQLQSYITQG----------------------------------
Rv2406c|M.tuberculosis_H37Rv ELEAEQQQLQSYITQG----------------------------------
MMAR_3726|M.marinum_M ELEAEQEQLQSYITQG----------------------------------
MUL_3669|M.ulcerans_Agy99 ELEAEQEQLQSYITQG----------------------------------
MAV_1776|M.avium_104 ELEAEQQQLQSYITQG----------------------------------
MSMEG_4564|M.smegmatis_MC2_155 ELETEQQQLQAYITQGR---------------------------------
MLBr_00387|M.leprae_Br4923 PLATKDNDGRLLVGAAVGVGGDAWVRAMMLVDAGVDVLIVDTAHAHNRLV
MAB_3721c|M.abscessus_ATCC_199 PNATKDSDGRLLVGAAVGVGEDAWTRAMSLVDAGVDVLIVDTAHAHNRKV
:.. : : .
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 LDMVGKLKVEIGDRVQVIGGNVATRSAAAALVEAGADAVKVGVGPGSTCT
MAB_3721c|M.abscessus_ATCC_199 LDMVGKLKAEVGERVDVVGGNVATRSAAAALVEAGADAVKVGVGPGSICT
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 TRVVAGVGAPQITAILEAVAACGPAGVPVIADGGLQYSGDIAKALAAGAS
MAB_3721c|M.abscessus_ATCC_199 TRVVAGVGAPQITAILEAATVCHPAGVPVIADGGMQYSGDIAKALAAGAS
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 TTMLGSLLAGTAEAPGELIFVNGKQFKSYRGMGSLGAMQGRGGDKSYSKD
MAB_3721c|M.abscessus_ATCC_199 TTMLGSLLAGTAESPGELIFVNGKQFKSYRGMGSMGAMQGRGATKSYSKD
Mflv_2670|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1455|M.thermoresistible__bu --------------------------------------------------
Mb2429c|M.bovis_AF2122/97 --------------------------------------------------
Rv2406c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3726|M.marinum_M --------------------------------------------------
MUL_3669|M.ulcerans_Agy99 --------------------------------------------------
MAV_1776|M.avium_104 --------------------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_00387|M.leprae_Br4923 RYFADDALSEDKLVPEGIEGRVPFRGPLSSVIHQLVGGLRAAMGYTGSPT
MAB_3721c|M.abscessus_ATCC_199 RYFQDDVLSEDKLVPEGIEGRVPFRGPLSTVMHQLTGGLRAAMGYTGASS
Mflv_2670|M.gilvum_PYR-GCK -------------------------------------
Mvan_3907|M.vanbaalenii_PYR-1 -------------------------------------
TH_1455|M.thermoresistible__bu -------------------------------------
Mb2429c|M.bovis_AF2122/97 -------------------------------------
Rv2406c|M.tuberculosis_H37Rv -------------------------------------
MMAR_3726|M.marinum_M -------------------------------------
MUL_3669|M.ulcerans_Agy99 -------------------------------------
MAV_1776|M.avium_104 -------------------------------------
MSMEG_4564|M.smegmatis_MC2_155 -------------------------------------
MLBr_00387|M.leprae_Br4923 IEVLQQAQFVRITPAGLKESHPHDVAMTVEAPNYYPR
MAB_3721c|M.abscessus_ATCC_199 IEELQRAQFVQITAAGLKESHPHDITMTAEAPNYYVR