For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TIVVGYLAGKVGPSALHLAVRVARMHKTSLTVATIVRRHWPTPSLARVDAEYELWSEQLAAASAREAQRY LRRLADGIEVSYHHRAHRSVSAGLLDVVEELEAEVLVLGSFPSGRRARVLIGSTADRLLHSSPVPVAITP RRYRCYTDRLTRLSCGYSATSGSVDVVRRCGHLASRYGVPMRVITFAVRGRTMYPPEVGLHAEASVLEAW AAQARELLEKLRINGVVSEDVVLQVVTGNGWAQALDAADWQDGEILALGTSPFGDVARVFLGSWSGKIIR YSPVPVLVLPG*
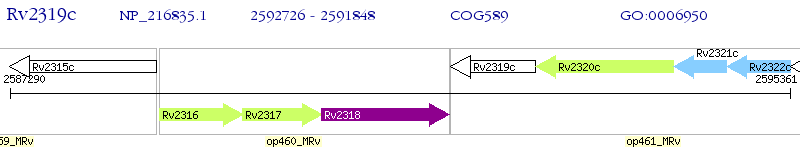
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2319c | - | - | 100% (292) | hypothetical protein Rv2319c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2346c | - | 1e-166 | 100.00% (292) | hypothetical protein Mb2346c |
| M. gilvum PYR-GCK | Mflv_1999 | - | 1e-44 | 33.56% (295) | UspA domain-containing protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3843 | - | 4e-85 | 53.92% (293) | hypothetical protein MAB_3843 |
| M. marinum M | MMAR_3620 | - | 1e-121 | 73.97% (292) | hypothetical protein MMAR_3620 |
| M. avium 104 | MAV_2507 | - | 3e-05 | 25.82% (306) | universal stress protein family protein |
| M. smegmatis MC2 155 | MSMEG_1411 | - | 2e-85 | 54.27% (293) | universal stress protein family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1245 | - | 1e-119 | 72.95% (292) | hypothetical protein MUL_1245 |
| M. vanbaalenii PYR-1 | Mvan_1573 | - | 1e-21 | 28.72% (296) | UspA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2319c|M.tuberculosis_H37Rv ---MTIVVGYLAGKVGPSALHLAVRVARMHKTSLTVATIVRRHWPTPSLA
Mb2346c|M.bovis_AF2122/97 ---MTIVVGYLAGKVGPSALHLAVRVARMHKTSLTVATIVRRHWPTPSLA
MMAR_3620|M.marinum_M ---MTVVVGYLAGRVGPSALHLAVRAARTLDTSLTVATILTKPWLTPPLA
MUL_1245|M.ulcerans_Agy99 ---MTVVVGYLAGRVGPSALHLAVRAARTLDTSLTVATILTKPWLTPPLA
MAB_3843|M.abscessus_ATCC_1997 ---MTVVVGYLSGKGGKGALHLAVESARVLETSLTVTTVVPKPWTTLSKA
MSMEG_1411|M.smegmatis_MC2_155 ---MTVVVGYLTGKAGASPLHLAVGAARTLQTSLAVATVVPRPWMTPSPA
Mflv_1999|M.gilvum_PYR-GCK ---MTIIAGFSASGQGPAPLHLAAQLARCTGERIVAVAIVERQWPQN---
Mvan_1573|M.vanbaalenii_PYR-1 ---MHLTVGYLATPTGDDGVALASALARTFDATVDVVIVVRHELPDG---
MAV_2507|M.avium_104 MAASSKRCGVLVGVDGSPASNFAVCWAARDAAMRNVPLTLVHMVNAATVW
* * :* * . : :
Rv2319c|M.tuberculosis_H37Rv RVDAEYELWSEQLAAASAREAQRYLRRLADG-IEVSYHHRAHRSVSAGLL
Mb2346c|M.bovis_AF2122/97 RVDAEYELWSEQLAAASAREAQRYLRRLADG-IEVSYHHRAHRSVSAGLL
MMAR_3620|M.marinum_M KG--DYSEWAEQVAEDSEKEAHRYLRTVATG-IEVSYCHREHRSVSGGLI
MUL_1245|M.ulcerans_Agy99 KG--DYSEWAEQVAEDSEKEAHRYLRTVATG-IEVSYCHREHRSVSGGLI
MAB_3843|M.abscessus_ATCC_1997 KIDAEYAQWAQKLSDDSKSEATAYLEKIGAG-IDVTFHNVAHRSVSGGLL
MSMEG_1411|M.smegmatis_MC2_155 RIDAEYAQYAAQLASTSAEQARQVISELDSE-LEVTFHKFAHRSVSDGLM
Mflv_1999|M.gilvum_PYR-GCK -ADPLEDEYLNYVGAQAQKSLKEMVDKLPGD-LDVWRMVHQAESVPSGLT
Mvan_1573|M.vanbaalenii_PYR-1 --HPGRAEYQQMLVNLGEQWAARAVAALAADGVTAESSAIVADSFAQALV
MAV_2507|M.avium_104 PQVPMAAEAVAWQEDDGRRVLQEAVKIAEDATRNGRNLAITTELWHAPPA
. :
Rv2319c|M.tuberculosis_H37Rv DVVEEL--EAEVLVLGSFPSGRRARVLIGSTADRLLHSSPVPVAITPRRY
Mb2346c|M.bovis_AF2122/97 DVVEEL--EAEVLVLGSFPSGRRARVLIGSTADRLLHSSPVPVAITPRRY
MMAR_3620|M.marinum_M EVVEEL--DAEMLILGSFPNGRRARVLIGSTADWVLHSSPVPVAISPRQY
MUL_1245|M.ulcerans_Agy99 EVVEEL--DAQMLILGSFPNGRRARVLIGSTADWVLHSSPVPVAISPRQY
MAB_3843|M.abscessus_ATCC_1997 EAVEAS--DADVLVVGSASEGRLGQVVLGSTGDRLLHSSPVPLAISPRGY
MSMEG_1411|M.smegmatis_MC2_155 EAVESL--NAEALVLGSAADGKLGQVVVGSTADRLLHSCPVPLVVSPRGY
Mflv_1999|M.gilvum_PYR-GCK ELAEEM--NADVVVVGSSSSGLLGRIALGSVTDRLVHTAAVPVAIAPRGY
Mvan_1573|M.vanbaalenii_PYR-1 EFAEEK--SSDLIVVGGARDGFFGRHTIGPVTGALLHSSSIPVALAPRGY
MAV_2507|M.avium_104 PTLAQLSEEAELVVVGSTGRGAIGRLLLGSVSSGLVRRARCPVAVIHDED
.:: :::*. * .: :*.. . ::: . *:.:
Rv2319c|M.tuberculosis_H37Rv R-CYTDRLTRLSCGYSATSGSVDVVRRCGHLASRYGVPMRVITFAVRGRT
Mb2346c|M.bovis_AF2122/97 R-CYTDRLTRLSCGYSATSGSVDVVRRCGHLASRYGVPMRVITFAVRGRT
MMAR_3620|M.marinum_M H-SRTDRLTRITCGYAATTDSVEVVRRCAELARQYGVPLRVITFAVRGRT
MUL_1245|M.ulcerans_Agy99 H-SRTDRLTRITCGYAATTDSVEVVRRCAELARQYGVPLRVITFAVRVRT
MAB_3843|M.abscessus_ATCC_1997 RGSRAGTVSRVTCGYPGTEDAVDVVQQAVRLTQRLHAPLRVVTFAVRGRT
MSMEG_1411|M.smegmatis_MC2_155 RRPRSGGLSRITVAYPGTRDAVDVVERIAALTQRLGVTMRVVTFAVRGRS
Mflv_1999|M.gilvum_PYR-GCK P-LQDSRIERLTVAYGGHADEGGLVAAGAELARRWSATLRIASFTVRPVT
Mvan_1573|M.vanbaalenii_PYR-1 AEDPDEHFLAVTAAVPSKPGDDNPLPFAIALAGAAGLRMRMLSLVSAEN-
MAV_2507|M.avium_104 PMMPYPQRAPVLVGIDGSPASELATAIAFDEALRRGVDLNAVHAWSDTQV
: . . : :.
Rv2319c|M.tuberculosis_H37Rv MYPPEVGLHAEASVLEAWAAQARE---LLEKLRINGVVSEDVVLQVVTGN
Mb2346c|M.bovis_AF2122/97 MYPPEVGLHAEASVLEAWAAQARE---LLEKLRINGVVSEDVVLQVVTGN
MMAR_3620|M.marinum_M MYPPEVGLHAEALVLEAWAEQARG---MLEMLQKDGIVEEDAVLEVITGN
MUL_1245|M.ulcerans_Agy99 MYPPEVGLHAEALVLEAWAEQARG---MLEMLQKDGIVEEDAVLEVITGN
MAB_3843|M.abscessus_ATCC_1997 MLTAGVGPEAEDAVLASWVTQSEE---ALSELRQAGIVTADVELKVVTGD
MSMEG_1411|M.smegmatis_MC2_155 MYPPLVGLSKEDLILQEWIEQAQD---SLRRLATDGVIGEDVEVVVASGS
Mflv_1999|M.gilvum_PYR-GCK MFSGAIETSAEDLVVQQWVRRTQEKIAAQLEAVRSGVDTLDAEVVIGAGA
Mvan_1573|M.vanbaalenii_PYR-1 ----LAEAGSAKEVRALQVAAAEEN---LAVAARALPDAPDIESLVADGM
MAV_2507|M.avium_104 FGLPGMDWPAVRSQAERSLAERLAG-------WQERYPDVTVHRMVVCDR
: .
Rv2319c|M.tuberculosis_H37Rv GWAQALDAADWQDGEILALGTSPFGDVARVFLGSWSGKIIRYSPVPVLVL
Mb2346c|M.bovis_AF2122/97 GWAQALDAADWQDGEILALGTSPFGDVARVFLGSWSGKIIRYSPVPVLVL
MMAR_3620|M.marinum_M GWQHALDAADWEDGEILAVGTSPRGDIARVFLGSNSGKIIRHSPVPVLVL
MUL_1245|M.ulcerans_Agy99 GWQHALDAADWEDGEILAVGTSPRGDIARVFLGSNSGRIIRHSPVPVLVL
MAB_3843|M.abscessus_ATCC_1997 TWDDALDNAEWNDGELLVLGSRPHSAVARVFLGSRATKIVRHSPVPVVVM
MSMEG_1411|M.smegmatis_MC2_155 AWDEALDSIEWIEGEVLAIGTSPAGAVRRVFLGERGTKILRYSPLPVLVL
Mflv_1999|M.gilvum_PYR-GCK TWRDAVEAIDWGSGDVLVLGSGAAGQVAQVFLGSAAARILRHSPVPTMIV
Mvan_1573|M.vanbaalenii_PYR-1 TLESALKKLRWDDGDLLVVGSSRFAAPRRIFLGSTAARILAGVDVPVIVV
MAV_2507|M.avium_104 PARQLIEQSES--AQLTVVGSHGRGGLAGTLLGSVSNAVVHSVRMPVIVA
:. .:: .:*: . :**. . :: :*.::
Rv2319c|M.tuberculosis_H37Rv PG----
Mb2346c|M.bovis_AF2122/97 PG----
MMAR_3620|M.marinum_M PG----
MUL_1245|M.ulcerans_Agy99 PG----
MAB_3843|M.abscessus_ATCC_1997 PG----
MSMEG_1411|M.smegmatis_MC2_155 PG----
Mflv_1999|M.gilvum_PYR-GCK PRREGL
Mvan_1573|M.vanbaalenii_PYR-1 PKAE--
MAV_2507|M.avium_104 RPS---