For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSPSRVSNTAVYAVLTIGAVITLSPFLLGLLTSFTSAHQFATGTPLQLPRPPTLANYADIADAGFRRAA VVTALMTAVILLGQLTFSVLAAYAFARLQFRGRDALFWVYVATLMVPGTVTVVPLYLMMAQLGLRNTFWA LVLPFMFGSPYAIFLLREHFRLIPDDLINAARLDGANTLDVIVHVVIPSSRPVLAALAMITVVSQWNNFM WPLVITSGHKWRVLTVATADLQSRFNDQWTLVMAATTVAIVPLIALFVTFQRHIVASIVVSGLK
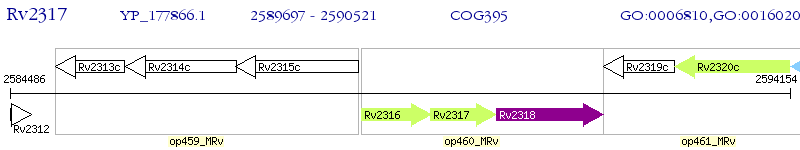
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2317 | uspB | - | 100% (274) | PROBABLE SUGAR-TRANSPORT INTEGRAL MEMBRANE PROTEIN ABC TRANSPORTER USPB |
| M. tuberculosis H37Rv | Rv2039c | - | 2e-47 | 37.69% (268) | sugar-transport integral membrane protein ABC transporter |
| M. tuberculosis H37Rv | Rv2834c | ugpE | 2e-39 | 31.05% (277) | sn-glycerol-3-phosphate transport integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2344 | uspB | 1e-153 | 100.00% (274) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3853 | - | 3e-29 | 29.89% (271) | binding-protein-dependent transport systems inner membrane |
| M. leprae Br4923 | MLBr_01769 | uspE | 1e-133 | 85.04% (274) | sugar transport integral membrane protein |
| M. abscessus ATCC 19977 | MAB_1716c | - | 1e-115 | 74.81% (266) | sugar ABC transporter, permease protein |
| M. marinum M | MMAR_3618 | uspB | 1e-133 | 86.62% (269) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_2089 | uspE | 1e-135 | 85.45% (275) | ABC transporter, permease protein UspE |
| M. smegmatis MC2 155 | MSMEG_4467 | - | 1e-117 | 79.47% (263) | ABC transporter, permease protein UspE |
| M. thermoresistible (build 8) | TH_2009 | - | 9e-25 | 29.01% (262) | sugar ABC transporter permease protein |
| M. ulcerans Agy99 | MUL_1247 | uspB | 1e-130 | 84.87% (271) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2542 | - | 5e-29 | 29.89% (271) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2317|M.tuberculosis_H37Rv -------------MSSPSR-----VSNTAVYAVLTIGAVITLSPFLLGLL
Mb2344|M.bovis_AF2122/97 -------------MSSPSR-----VSNTAVYAVLTIGAVITLSPFLLGLL
MMAR_3618|M.marinum_M -------------MTSLSRRLSRRVPTTVIYAGLVLGALITLLPFALGLL
MUL_1247|M.ulcerans_Agy99 ------------------------MPTRVIYAGLVLGALITLLPFALGLL
MLBr_01769|M.leprae_Br4923 -------------MSSPSR-----LSNAVTYAGLTLGALITVAPFALGLL
MAV_2089|M.avium_104 -------------MTSPDR----ARANIAIYAGLLVGALITLAPFTLGLL
MSMEG_4467|M.smegmatis_MC2_155 ----------------------------MIYAGLLLGAVITLLPFGLGLL
MAB_1716c|M.abscessus_ATCC_199 --------------MTWSR-------NVLVYLLLTVGAVLTLGPFLLAVS
Mflv_3853|M.gilvum_PYR-GCK ----------------------MTKNTKWWTIAGIVIVIYSFVPMLWMIS
Mvan_2542|M.vanbaalenii_PYR-1 ----------------------MTKNTKWWTIAGIVIVIYSFVPMLWMIS
TH_2009|M.thermoresistible__bu MSTATVTTTAPVPTEPSKAAKVKRRSFDPWGVVAWIAAIGFFFPVFWMVL
: .: . *. :
Rv2317|M.tuberculosis_H37Rv TSFTSAHQFATGTPLQLPRPPTLANYADIAD-AGFRRAAVVTALMTAVIL
Mb2344|M.bovis_AF2122/97 TSFTSAHQFATGTPLQLPRPPTLANYADIAD-AGFRRAAVVTALMTAVIL
MMAR_3618|M.marinum_M TSFTSAHQFATGTPLQLPHPPTLANYADLGG-AGFGRAALVTALMTAVIL
MUL_1247|M.ulcerans_Agy99 TSFTSAHQFATGTPLQLPHPPTLTNYADLGG-AGFGRAALVTALMTTVIL
MLBr_01769|M.leprae_Br4923 TSFTSAHQFTTGTPLQLPRPPTLANYAGLSG-AGFLRATAVTALMTAVIL
MAV_2089|M.avium_104 TAFTSAHQFVTGTPLQLPRPPTLSNFADLAG-AGFGRAGAVTALMTAVIL
MSMEG_4467|M.smegmatis_MC2_155 TSFTSAQQFVTESPLSLPRPPTLANYLGLAD-AGFGRAIAVTALMTAVIL
MAB_1716c|M.abscessus_ATCC_199 ASLKTAKQFATGTPLAPPNPFTLANYTGLAD-AGFGRAALVTVLMTAIIV
Mflv_3853|M.gilvum_PYR-GCK LAFKPPSDIVSGEPSFLPSTVTFDNFAQIFSNPLFTRALINSIGIALVAT
Mvan_2542|M.vanbaalenii_PYR-1 LAFKPPSDIVSGKPSFLPSSITFDNFAQIFSQPLFTRALINSIGIAVIAT
TH_2009|M.thermoresistible__bu TAFKQEVDAYTTTPKVFFAP-TLDQFRAVFD-QGIGTALLNSLFATVVST
::. : : * . *: :: : . : * : : :
Rv2317|M.tuberculosis_H37Rv LGQLTFSVLAAYAFARLQFRGRDALFWVYVATLMVPGTVTVVPLYLMMAQ
Mb2344|M.bovis_AF2122/97 LGQLTFSVLAAYAFARLQFRGRDALFWVYVATLMVPGTVTVVPLYLMMAQ
MMAR_3618|M.marinum_M LGQMTFSVLAAYAFARLEFPGRDGLFWVYIATLMVPATVTVVPLYLMMAQ
MUL_1247|M.ulcerans_Agy99 LGQMTFSVLAAYAFARLEFPGRDGLFWVYIATLMVPATVTVVPLYLMMAQ
MLBr_01769|M.leprae_Br4923 LGQLTFSVLAGYAFARLRFPGRDTLFWAYIMTLMVPATVTVVPMYMMMAQ
MAV_2089|M.avium_104 VGQLTFSVLAGYAFARLRFPGRDALFWVYIATLMVPGTVTIVPMYLMMAQ
MSMEG_4467|M.smegmatis_MC2_155 LGQLVFSVLAAYAFARLEFPGRDVLFWVYLATLMVPATVTVVPLYLIMAE
MAB_1716c|M.abscessus_ATCC_199 IGQLTFSVLAAYAFARLRFPGRDGLFWIYLATLMIPATVTVVPMYLMLTQ
Mflv_3853|M.gilvum_PYR-GCK VISVIIAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRG
Mvan_2542|M.vanbaalenii_PYR-1 VISVIIAMFAAYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRG
TH_2009|M.thermoresistible__bu LLVLALGVPAAFALSLRPVRKTQDALFFFMSTKMLPIVAAIIPLYVIVSN
: : :.: *.:*:: . . : : *.* .. : *:: :
Rv2317|M.tuberculosis_H37Rv LGLRNTFWALVLPFM-FGSPYAIFLLREHFRLIPDDLINAARLDGANTLD
Mb2344|M.bovis_AF2122/97 LGLRNTFWALVLPFM-FGSPYAIFLLREHFRLIPDDLINAARLDGANTLD
MMAR_3618|M.marinum_M LGLRNTFWALVLPFL-FGSPYAIFLLREHFRIIPNDMINAARLDGANTLD
MUL_1247|M.ulcerans_Agy99 LGLRNTFWALVLPFL-FGSPYAIFLLREHFRIIPNDMINAARLDGANTLD
MLBr_01769|M.leprae_Br4923 VGLRNTFWALVLPFM-FGSPYAIFLLREHFRIIPNDLINAARLDGANTLD
MAV_2089|M.avium_104 LGLRNTFWALVLPFM-FGSPYAIFLLREHFRMIPNDLVNAARLDGAHTLD
MSMEG_4467|M.smegmatis_MC2_155 AGLRNTFWALVLPFV-FGSPYAIFLLREYFRGIPGDLINAARLDGANTLD
MAB_1716c|M.abscessus_ATCC_199 VGLRNTFWALVIPFM-FGSPYAIFLLREHFRTIPDDLIRAARLDGANTLD
Mflv_3853|M.gilvum_PYR-GCK LGIYDTWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQ
Mvan_2542|M.vanbaalenii_PYR-1 LGIYDTWLGLIIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQ
TH_2009|M.thermoresistible__bu VGLLDNIWALIILYTAMNLPIAVWMMRSFFLEVPGELLEAASLDGAGTWR
*: :. .*:: : : * ::: : .* :* :: .** :***
Rv2317|M.tuberculosis_H37Rv VIVHVVIPSSRPVLAALAMITVVSQWNNFMWPLVITSGHKWRVLTVATAD
Mb2344|M.bovis_AF2122/97 VIVHVVIPSSRPVLAALAMITVVSQWNNFMWPLVITSGHKWRVLTVATAD
MMAR_3618|M.marinum_M LLVHVVIPSSRPVLAAMTLITVVSQWNNFMWPLVITSGHKWRVLTVATAD
MUL_1247|M.ulcerans_Agy99 VLVHVVIPSNRPVLAAMTLITVVSQWNNFMWPLVITSGHKWRVLTVATAD
MLBr_01769|M.leprae_Br4923 VIVHVVIPSNRSVLAALTLITVVSQWNNFMWPLVITSGNKWRVLTVATAD
MAV_2089|M.avium_104 VIVHVVIPSSRPVLAALALITVVSQWNNFMWPLVITSGHKWRVLTVATAD
MSMEG_4467|M.smegmatis_MC2_155 IITHVVVPASRPILVTLAMITVVSQWNNFLWPLVISSGDTWRVLTVATAG
MAB_1716c|M.abscessus_ATCC_199 ILWHVVVPVSRPILITLTLITVVSQWNNFMWPLVITSGGNWRVLTVATAG
Mflv_3853|M.gilvum_PYR-GCK AFRKVIVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAF
Mvan_2542|M.vanbaalenii_PYR-1 AFRKVIVPLAAPGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAF
TH_2009|M.thermoresistible__bu TMREVILPIVSPGIAATALICVIFAWNEFFFAVNLTAVQAQTMPVFLVGF
: .*::* . : : ::: .. **:*::.: ::: .
Rv2317|M.tuberculosis_H37Rv LQ--SRFNDQWTLVMAATTVAIVPLIALFVTFQRHIVASIVVSGLK-
Mb2344|M.bovis_AF2122/97 LQ--SRFNDQWTLVMAATTVAIVPLIALFVTFQRHIVASIVVSGLK-
MMAR_3618|M.marinum_M LQ--SRFNAQWTLVMAATTVAIVPLIVLFVAAQRHIVSSIVVSGLK-
MUL_1247|M.ulcerans_Agy99 LQ--SRFNAQWTLVMAATTVAIVPLIVLFVAAQRHIVSSIAVSGLK-
MLBr_01769|M.leprae_Br4923 LQ--SRFNAQWTLVMAATTVAIVPLIVLFVGFQRHIIPSIVVSGLK-
MAV_2089|M.avium_104 LQ--TRFNAQWTLVMAATTIAIVPLVTLFVIFQRHIVASIVVSGLK-
MSMEG_4467|M.smegmatis_MC2_155 LQ--TQYNAQWTLVMAATTVAIVPLIVVFATLSRHIVRSIVVTGIK-
MAB_1716c|M.abscessus_ATCC_199 LQ--SQFNAQWPIVMAATTVAIVPLVILFVAFQKYIVRSITLSGLK-
Mflv_3853|M.gilvum_PYR-GCK FQGASYFESPVPYIMAASVIVTIPVVILVLVFQRRIVAGLTSGAVKG
Mvan_2542|M.vanbaalenii_PYR-1 FQGASYFESPVPYIMAASVIVTIPVVILVLIFQRRIVAGLTSGAVKG
TH_2009|M.thermoresistible__bu IAG---EGLYWAKLCAASTMAALPVILAGWLAQNKLVRGLSFGAIK-
: . : **:.:. :*:: .. :: .: .:*