For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSPSRLSNAVTYAGLTLGALITVAPFALGLLTSFTSAHQFTTGTPLQLPRPPTLANYAGLSGAGFLRAT AVTALMTAVILLGQLTFSVLAGYAFARLRFPGRDTLFWAYIMTLMVPATVTVVPMYMMMAQVGLRNTFWA LVLPFMFGSPYAIFLLREHFRIIPNDLINAARLDGANTLDVIVHVVIPSNRSVLAALTLITVVSQWNNFM WPLVITSGNKWRVLTVATADLQSRFNAQWTLVMAATTVAIVPLIVLFVGFQRHIIPSIVVSGLK
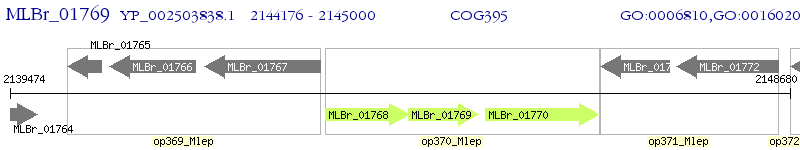
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01769 | uspE | - | 100% (274) | sugar transport integral membrane protein |
| M. leprae Br4923 | MLBr_01425 | - | 4e-44 | 34.33% (268) | putative ABC-transport protein, inner membrane component |
| M. leprae Br4923 | MLBr_01088 | - | 6e-23 | 28.45% (232) | putative ABC-transport protein, inner membrane component |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2344 | uspB | 1e-132 | 85.04% (274) | sugar-transport integral membrane protein ABC transporter |
| M. gilvum PYR-GCK | Mflv_3853 | - | 6e-26 | 26.97% (267) | binding-protein-dependent transport systems inner membrane |
| M. tuberculosis H37Rv | Rv2317 | uspB | 1e-132 | 85.04% (274) | sugar-transport integral membrane protein ABC transporter |
| M. abscessus ATCC 19977 | MAB_1716c | - | 1e-112 | 72.93% (266) | sugar ABC transporter, permease protein |
| M. marinum M | MMAR_3618 | uspB | 1e-132 | 86.62% (269) | sugar-transport integral membrane protein ABC transporter |
| M. avium 104 | MAV_2089 | uspE | 1e-132 | 84.73% (275) | ABC transporter, permease protein UspE |
| M. smegmatis MC2 155 | MSMEG_4467 | - | 1e-117 | 77.27% (264) | ABC transporter, permease protein UspE |
| M. thermoresistible (build 8) | TH_2867 | - | 1e-21 | 28.39% (236) | PUTATIVE binding-protein-dependent transport systems inner |
| M. ulcerans Agy99 | MUL_1247 | uspB | 1e-131 | 86.57% (268) | sugar-transport integral membrane protein ABC transporter |
| M. vanbaalenii PYR-1 | Mvan_2542 | - | 1e-25 | 26.22% (267) | binding-protein-dependent transport systems inner membrane |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2344|M.bovis_AF2122/97 MSSPSR-----VSNTAVYAVLTIGAVITLSPFLLGLLTSFTSAHQFATGT
Rv2317|M.tuberculosis_H37Rv MSSPSR-----VSNTAVYAVLTIGAVITLSPFLLGLLTSFTSAHQFATGT
MMAR_3618|M.marinum_M MTSLSRRLSRRVPTTVIYAGLVLGALITLLPFALGLLTSFTSAHQFATGT
MUL_1247|M.ulcerans_Agy99 -----------MPTRVIYAGLVLGALITLLPFALGLLTSFTSAHQFATGT
MLBr_01769|M.leprae_Br4923 MSSPSR-----LSNAVTYAGLTLGALITVAPFALGLLTSFTSAHQFTTGT
MAV_2089|M.avium_104 MTSPDR----ARANIAIYAGLLVGALITLAPFTLGLLTAFTSAHQFVTGT
MSMEG_4467|M.smegmatis_MC2_155 ---------------MIYAGLLLGAVITLLPFGLGLLTSFTSAQQFVTES
MAB_1716c|M.abscessus_ATCC_199 -MTWSR-------NVLVYLLLTVGAVLTLGPFLLAVSASLKTAKQFATGT
Mflv_3853|M.gilvum_PYR-GCK ---------MTKNTKWWTIAGIVIVIYSFVPMLWMISLAFKPPSDIVSGE
Mvan_2542|M.vanbaalenii_PYR-1 ---------MTKNTKWWTIAGIVIVIYSFVPMLWMISLAFKPPSDIVSGK
TH_2867|M.thermoresistible__bu ----VNERVTARRATGWIVIDLLVILYALFPVLWILSLSLKSTATVKDGK
: : :. *. : ::... .
Mb2344|M.bovis_AF2122/97 PLQLPRPPTLANYADIADAG----FRRAAVVTALMTAVILLGQLTFSVLA
Rv2317|M.tuberculosis_H37Rv PLQLPRPPTLANYADIADAG----FRRAAVVTALMTAVILLGQLTFSVLA
MMAR_3618|M.marinum_M PLQLPHPPTLANYADLGGAG----FGRAALVTALMTAVILLGQMTFSVLA
MUL_1247|M.ulcerans_Agy99 PLQLPHPPTLTNYADLGGAG----FGRAALVTALMTTVILLGQMTFSVLA
MLBr_01769|M.leprae_Br4923 PLQLPRPPTLANYAGLSGAG----FLRATAVTALMTAVILLGQLTFSVLA
MAV_2089|M.avium_104 PLQLPRPPTLSNFADLAGAG----FGRAGAVTALMTAVILVGQLTFSVLA
MSMEG_4467|M.smegmatis_MC2_155 PLSLPRPPTLANYLGLADAG----FGRAIAVTALMTAVILLGQLVFSVLA
MAB_1716c|M.abscessus_ATCC_199 PLAPPNPFTLANYTGLADAG----FGRAALVTVLMTAIIVIGQLTFSVLA
Mflv_3853|M.gilvum_PYR-GCK PSFLPSTVTFDNFAQIFS---NPLFTRALINSIGIALVATVISVIIAMFA
Mvan_2542|M.vanbaalenii_PYR-1 PSFLPSSITFDNFAQIFS---QPLFTRALINSIGIAVIATVISVIIAMFA
TH_2867|M.thermoresistible__bu --LIPDEITFDNYKAIFRGGGSGVFTSALINSIGIGLITTVIAVVVGGMA
* *: *: : * * : : : : : .. :*
Mb2344|M.bovis_AF2122/97 AYAFARLQFRGRDALFWVYVATLMVPGTVTVVPLYLMMAQLGLRNTFWAL
Rv2317|M.tuberculosis_H37Rv AYAFARLQFRGRDALFWVYVATLMVPGTVTVVPLYLMMAQLGLRNTFWAL
MMAR_3618|M.marinum_M AYAFARLEFPGRDGLFWVYIATLMVPATVTVVPLYLMMAQLGLRNTFWAL
MUL_1247|M.ulcerans_Agy99 AYAFARLEFPGRDGLFWVYIATLMVPATVTVVPLYLMMAQLGLRNTFWAL
MLBr_01769|M.leprae_Br4923 GYAFARLRFPGRDTLFWAYIMTLMVPATVTVVPMYMMMAQVGLRNTFWAL
MAV_2089|M.avium_104 GYAFARLRFPGRDALFWVYIATLMVPGTVTIVPMYLMMAQLGLRNTFWAL
MSMEG_4467|M.smegmatis_MC2_155 AYAFARLEFPGRDVLFWVYLATLMVPATVTVVPLYLIMAEAGLRNTFWAL
MAB_1716c|M.abscessus_ATCC_199 AYAFARLRFPGRDGLFWIYLATLMIPATVTVVPMYLMLTQVGLRNTFWAL
Mflv_3853|M.gilvum_PYR-GCK AYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRGLGIYDTWLGL
Mvan_2542|M.vanbaalenii_PYR-1 AYAIARLEFPGKKVLLSLALGIAMFPQAALVGPLFDMWRGLGIYDTWLGL
TH_2867|M.thermoresistible__bu AYAVARLNFPGKRLLIGVALLIAMFPQISLVTPLFNIERQLGLFDTWAGL
.**.***.* *: *: : *.* : *:: : *: :*: .*
Mb2344|M.bovis_AF2122/97 VLPFM-FGSPYAIFLLREHFRLIPDDLINAARLDGANTLDVIVHVVIPSS
Rv2317|M.tuberculosis_H37Rv VLPFM-FGSPYAIFLLREHFRLIPDDLINAARLDGANTLDVIVHVVIPSS
MMAR_3618|M.marinum_M VLPFL-FGSPYAIFLLREHFRIIPNDMINAARLDGANTLDLLVHVVIPSS
MUL_1247|M.ulcerans_Agy99 VLPFL-FGSPYAIFLLREHFRIIPNDMINAARLDGANTLDVLVHVVIPSN
MLBr_01769|M.leprae_Br4923 VLPFM-FGSPYAIFLLREHFRIIPNDLINAARLDGANTLDVIVHVVIPSN
MAV_2089|M.avium_104 VLPFM-FGSPYAIFLLREHFRMIPNDLVNAARLDGAHTLDVIVHVVIPSS
MSMEG_4467|M.smegmatis_MC2_155 VLPFV-FGSPYAIFLLREYFRGIPGDLINAARLDGANTLDIITHVVVPAS
MAB_1716c|M.abscessus_ATCC_199 VIPFM-FGSPYAIFLLREHFRTIPDDLIRAARLDGANTLDILWHVVVPVS
Mflv_3853|M.gilvum_PYR-GCK IIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQAFRKVIVPLA
Mvan_2542|M.vanbaalenii_PYR-1 IIPYLTFALPLSIWTMSAFFRQIPWEMEHAAQVDGATQWQAFRKVIVPLA
TH_2867|M.thermoresistible__bu IIPYITFALPLAIYTLSAFFREIPWDLEKAAKMDGATPGQAFRKVIAPLA
::*:: *. * :*: : .** ** :: .**::*** : : :*: *
Mb2344|M.bovis_AF2122/97 RPVLAALAMITVVSQWNNFMWPLVITSGHKWRVLTVATADLQ--SRFNDQ
Rv2317|M.tuberculosis_H37Rv RPVLAALAMITVVSQWNNFMWPLVITSGHKWRVLTVATADLQ--SRFNDQ
MMAR_3618|M.marinum_M RPVLAAMTLITVVSQWNNFMWPLVITSGHKWRVLTVATADLQ--SRFNAQ
MUL_1247|M.ulcerans_Agy99 RPVLAAMTLITVVSQWNNFMWPLVITSGHKWRVLTVATADLQ--SRFNAQ
MLBr_01769|M.leprae_Br4923 RSVLAALTLITVVSQWNNFMWPLVITSGNKWRVLTVATADLQ--SRFNAQ
MAV_2089|M.avium_104 RPVLAALALITVVSQWNNFMWPLVITSGHKWRVLTVATADLQ--TRFNAQ
MSMEG_4467|M.smegmatis_MC2_155 RPILVTLAMITVVSQWNNFLWPLVISSGDTWRVLTVATAGLQ--TQYNAQ
MAB_1716c|M.abscessus_ATCC_199 RPILITLTLITVVSQWNNFMWPLVITSGGNWRVLTVATAGLQ--SQFNAQ
Mflv_3853|M.gilvum_PYR-GCK APGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAFFQGASYFESP
Mvan_2542|M.vanbaalenii_PYR-1 APGVFTTAILTFFFCWNDFLFAISLTSTDSARTVPAALAFFQGASYFESP
TH_2867|M.thermoresistible__bu APGIVTAAILVFIFAWNDLLLALSLTATERAITAPVAIANFTGSSQFEEP
. : : :::... **::: .: ::: . ..* * : : ::
Mb2344|M.bovis_AF2122/97 WTLVMAATTVAIVPLIALFVTFQRHIVASIVVSGLK-
Rv2317|M.tuberculosis_H37Rv WTLVMAATTVAIVPLIALFVTFQRHIVASIVVSGLK-
MMAR_3618|M.marinum_M WTLVMAATTVAIVPLIVLFVAAQRHIVSSIVVSGLK-
MUL_1247|M.ulcerans_Agy99 WTLVMAATTVAIVPLIVLFVAAQRHIVSSIAVSGLK-
MLBr_01769|M.leprae_Br4923 WTLVMAATTVAIVPLIVLFVGFQRHIIPSIVVSGLK-
MAV_2089|M.avium_104 WTLVMAATTIAIVPLVTLFVIFQRHIVASIVVSGLK-
MSMEG_4467|M.smegmatis_MC2_155 WTLVMAATTVAIVPLIVVFATLSRHIVRSIVVTGIK-
MAB_1716c|M.abscessus_ATCC_199 WPIVMAATTVAIVPLVILFVAFQKYIVRSITLSGLK-
Mflv_3853|M.gilvum_PYR-GCK VPYIMAASVIVTIPVVILVLVFQRRIVAGLTSGAVKG
Mvan_2542|M.vanbaalenii_PYR-1 VPYIMAASVIVTIPVVILVLIFQRRIVAGLTSGAVKG
TH_2867|M.thermoresistible__bu TGSIAAGAMVITIPIIVFVLIFQRRIVSGLTSGAVKG
: *.: : :*:: .. .: *: .:. .:*