For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAGSDLDDFRGLLAKAFDERVVAWTAEAEAQERFPRQLIEHLGVCGVFDAKWATDARPDVGKLVELAFA LGQLASAGIGVGVSLHDSAIAILRRFGKSDYLRDICDQAIRGAAVLCIGASEESGGSDLQIVETEIRSRD GGFEVRGVKKFVSLSPIADHIMVVARSVDHDPTSRHGNVAVVAVPAAQVSVQTPYRKVGAGPLDTAAVCI DTWVPADALVARAGTGLAAISWGLAHERMSIAGQIAASCQRAIGITLARMMSRRQFGQTLFEHQALRLRM ADLQARVDLLRYALHGIAEQGRLELRTAAAVKVTAARLGEEVISECMHIFGGAGYLVDETTLGKWWRDMK LARVGGGTDEVLWELVAAGMTPDHDGYAAVVGASKA
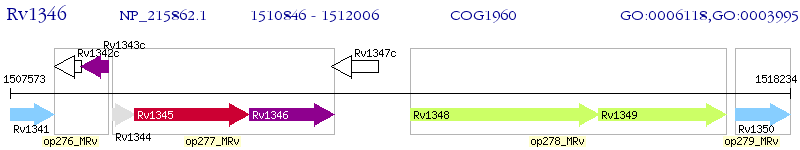
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1346 | fadE14 | - | 100% (386) | POSSIBLE ACYL-CoA DEHYDROGENASE FADE14 |
| M. tuberculosis H37Rv | Rv0975c | fadE13 | 1e-27 | 26.52% (362) | acyl-CoA dehydrogenase FADE13 |
| M. tuberculosis H37Rv | Rv2500c | fadE19 | 5e-22 | 23.28% (378) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1381 | fadE14 | 0.0 | 100.00% (386) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1884 | - | 6e-30 | 28.34% (374) | acyl-CoA dehydrogenase domain-containing protein |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 9e-20 | 22.53% (364) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1851 | - | 2e-30 | 27.78% (378) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4532 | fadE13 | 3e-27 | 27.35% (362) | acyl-CoA dehydrogenase FadE13 |
| M. avium 104 | MAV_2876 | - | 0.0 | 82.46% (382) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2130 | - | 1e-174 | 79.52% (376) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4231 | - | 1e-176 | 79.22% (385) | PUTATIVE putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_4704 | fadE13 | 8e-27 | 26.80% (362) | acyl-CoA dehydrogenase FadE13 |
| M. vanbaalenii PYR-1 | Mvan_1944 | - | 1e-174 | 79.11% (383) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1346|M.tuberculosis_H37Rv ---------------MTAG--------SDLDDFRGLLAKAFDERVVAWTA
Mb1381|M.bovis_AF2122/97 ---------------MTAG--------SDLDDFRGLLAKAFDERVVAWTA
MAV_2876|M.avium_104 ------------MTTLSAAT------TADIDHYRTVLAGAFDDQVLEWTR
MSMEG_2130|M.smegmatis_MC2_155 ---------------MTASAPALTPSTTTVEEYRELLDRVFDDQVTQWTA
TH_4231|M.thermoresistible__bu ---------------MTAVS------ETSVDDYRDLLGRVFDERVTAWTA
Mvan_1944|M.vanbaalenii_PYR-1 ---------------MTVSM------DVDIDGYRELLSEVFDEQVTAWTA
MMAR_4532|M.marinum_M ---------MSIWNTP-----------ERE-ELRKTVRSFAEREVLPHID
MUL_4704|M.ulcerans_Agy99 ---------MSIWNTP-----------ERE-ELRKSVRSFAEREVLPHID
Mflv_1884|M.gilvum_PYR-GCK ---------MSIWTTP-----------ERD-DLRRTVRAFAEREVLPFAD
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLP-----------EEHNELRATIRALAEKEIAPHAA
MAB_1851|M.abscessus_ATCC_1997 ------------MTFS-----------SEHDDFRRTIRRYIEEKINPRVD
* : : .:
Rv1346|M.tuberculosis_H37Rv EAEAQERFP-RQLIEHLGVCGVFDAKWATD---ARPDVGKLVELAFALGQ
Mb1381|M.bovis_AF2122/97 EAEAQERFP-RQLIEHLGVCGVFDAKWATD---ARPDVGKLVELAFALGQ
MAV_2876|M.avium_104 EAEARQRFP-RELIEHLGARGVFSEKWCGG---MLPDVGKLVELARALGR
MSMEG_2130|M.smegmatis_MC2_155 EAEETERFP-RQLIEHLGRNGVFTEKWGDA---QQPDVAKLIELAFSLGR
TH_4231|M.thermoresistible__bu EAEETERFP-RALIEYLGRSGVFAEKWQDH---QLPDVAKLIELALALGR
Mvan_1944|M.vanbaalenii_PYR-1 EAVASERFP-RKLIEHLGAAGVFRSKWGPAGG-QHPDVAKLNELAFRLGH
MMAR_4532|M.marinum_M EWERTGELP-RDLHRRAGAAGLLGAGLPESAGGGGGDGADSVIICEQMHQ
MUL_4704|M.ulcerans_Agy99 EWERTGELP-LDLHRRAGAAGLLGAGLPESAGGGGGDGADSVIICEQMHQ
Mflv_1884|M.gilvum_PYR-GCK EWERAREIP-RDLHRAAAAAGLLGAGFPEEVGGDGGDGADAVVICEEMHE
MLBr_00737|M.leprae_Br4923 DVDQRARFP-EEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVAR
MAB_1851|M.abscessus_ATCC_1997 EWEREQMMPLHEVIADMAKLGFVGLEYDPEYGGQGADHLFTLVLAEELGR
: :* *. * : : .
Rv1346|M.tuberculosis_H37Rv LASAG-IGVGVSLHDSAIAILRRFGKSDYLRDICDQAIRGAAVLCIGASE
Mb1381|M.bovis_AF2122/97 LASAG-IGVGVSLHDSAIAILRRFGKSDYLRDICDQAIRGAAVLCIGASE
MAV_2876|M.avium_104 LSSAG-IGVGVSLHDSAIAVLRRFGKSDYLRDICERAIAGQAVLCIGASE
MSMEG_2130|M.smegmatis_MC2_155 LGSAG-IGVGVSLHDSAICVLRRFGRSDYLRNICEQAIRGEAVLCIGASE
TH_4231|M.thermoresistible__bu LGSAG-IGVGVSLHDSAIAILRRFGRSDHLREIAEQAIRGEAVLCIGASE
Mvan_1944|M.vanbaalenii_PYR-1 LGSAG-IGVGVSLHDSAIAVLRRFGRSDHLKAVCEQAIDGEAVLCIGASE
MMAR_4532|M.marinum_M SGAPGGVFASLFTCGIAVPHMIASGDERLIEEFVRPTLAGDKIGALAITE
MUL_4704|M.ulcerans_Agy99 SGAPGGVFASLFTCGIAVPHMIASGDERLIEEFVRPTLAGNKIGALAITE
Mflv_1884|M.gilvum_PYR-GCK SGCPGGVFASLFTSGIAVPHMIASQDERLIDTYVRPTLRGENIGSLAITE
MLBr_00737|M.leprae_Br4923 VDASASLIPAVNKLGTMG--LILRGSEELKKQVLPSLAAEGAMASYALSE
MAB_1851|M.abscessus_ATCC_1997 VDHGSFPMAFGVHVAMATPSLHKHGSDELKREFLAPALRGEQIAAIAVTE
. : . : . . :*
Rv1346|M.tuberculosis_H37Rv ESGGSDLQIVETEIRSRDGGFEVRGVKKFVSLSPIADHIMVVARSVDHDP
Mb1381|M.bovis_AF2122/97 ESGGSDLQIVETEIRSRDGGFEVRGVKKFVSLSPIADHIMVVARSVDHDP
MAV_2876|M.avium_104 ESGGSDLQIVRTEMSSRDGGFDVRGVKKFVSLSPIADHIMVVARSIDHDS
MSMEG_2130|M.smegmatis_MC2_155 ESGGSDLQIVETEVRSARGGFEVKGIKKFVSLSPIADHIIVVARGVDHDP
TH_4231|M.thermoresistible__bu ESGGSDLQIVETEVRSEGDGFRVRGTKKFVSLSPIADHIVVVARSVDHDP
Mvan_1944|M.vanbaalenii_PYR-1 ESGGSDLQIVETEVRSAREGFEIRGIKKFVSLSPIADHIIVVARGVDHDP
MMAR_4532|M.marinum_M PGGGSDVGHLRTSAVRDGDHFIVNGAKTYITSGVR-ADFVVTAVRTGGPG
MUL_4704|M.ulcerans_Agy99 PGGGSDVGHLRTSAVREGDHFIVNGAKTYITSGVR-ADFVVTAVRTGGPG
Mflv_1884|M.gilvum_PYR-GCK PGGGSDVGHLTTRAVRDGDCFIVNGAKTYITSGVR-ADYVVTAARTGGPG
MLBr_00737|M.leprae_Br4923 REAGSDAASMRTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKG
MAB_1851|M.abscessus_ATCC_1997 PDAGSDVAGIKTHARRDGDDWVINGSKMFITNSVQADWLCVLARTSDEGG
.*** : * : :.* * ::: . * *
Rv1346|M.tuberculosis_H37Rv TSRHGNVAVVAVPAAQVSVQTPYRKVGAGPLDTAAVCID-TWVPADALVA
Mb1381|M.bovis_AF2122/97 TSRHGNVAVVAVPAAQVSVQTPYRKVGAGPLDTAAVCID-TWVPADALVA
MAV_2876|M.avium_104 ASKHGNVALIAVPTSQVSVQRPYAKVGAGPLDTAAVHID-TWVPADALVA
MSMEG_2130|M.smegmatis_MC2_155 TSRHGNVLVIAVPTSQCEVQTPYRKVGAGPLDTAAVHID-TWVPEQALVA
TH_4231|M.thermoresistible__bu SSRHGNVMLIAVPTTQVEVQTPYRKVGAGPLDTAAVHID-TWVPGDALIA
Mvan_1944|M.vanbaalenii_PYR-1 TSRHGNVLVVAVPTRQVEIQTPYRKVGAGPLDTAAVHID-TWVPADHLVA
MMAR_4532|M.marinum_M AGGVS-LLVVEKGTPGFVVSRKLDKMGWRSSDTAELSYTDARVPAANLVG
MUL_4704|M.ulcerans_Agy99 AGGVS-LLVVEKGTPGFVVSRKLDKMGWRSSDTAELSYTDARIPAANLVG
Mflv_1884|M.gilvum_PYR-GCK AAGIS-LIVVDKGTPGFAVSRKLDKMGWRSSDTAELSYTDVRVPAANVVG
MLBr_00737|M.leprae_Br4923 ANGIS-AFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIG
MAB_1851|M.abscessus_ATCC_1997 YAGMS-QIIVPTKAPGYEVR-KLDKLGMHASDTGLITFDDVRVPVSNTIG
. :: : *:* * : :* :.
Rv1346|M.tuberculosis_H37Rv RAGTGLAAISWGLAHERMSIAGQIAASCQRAIGITLARMMSRRQFGQTLF
Mb1381|M.bovis_AF2122/97 RAGTGLAAISWGLAHERMSIAGQIAASCQRAIGITLARMMSRRQFGQTLF
MAV_2876|M.avium_104 RAGTGLAAISWGLAHERMSIAGQIAASCQRAIGITLARMMTRRQFGRTLF
MSMEG_2130|M.smegmatis_MC2_155 RAGTGLAAISWGLAHERMSIAGQVASGSQRILGITLARMMKRRQFGHTLY
TH_4231|M.thermoresistible__bu RPGTGLAAISWGLAHERLSIAGQVAAGCQRVIGITLARMMKRRQFGSTLY
Mvan_1944|M.vanbaalenii_PYR-1 RAGTGLAAISWGLAHERMSIAGQVASSCQRVLGVTHARMVQRRQFGATLF
MMAR_4532|M.marinum_M AENSGFAQIAQAFVSERIALAAQAYSSAQRCLDITVQWCRDRETFGRPLI
MUL_4704|M.ulcerans_Agy99 AENSGFAQIAQAFVSERIALAAQAYSSAQRCLDITVQWCRDRETFGRPLI
Mflv_1884|M.gilvum_PYR-GCK AENTGFLQIAAAFVSERVGLAAQAYSSAQRCLDLTVEWCRNRQTFGKPLI
MLBr_00737|M.leprae_Br4923 EPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESIS
MAB_1851|M.abscessus_ATCC_1997 EIGRGFQQQMSQFVMERMFGAYGIPASVNRALQRTKEYALARHVFGKPLT
. *: : * . . : : : *. ** .:
Rv1346|M.tuberculosis_H37Rv EHQALRLRMADLQARVDLLRYALHGIAEQGRLELR----TAAAVKVTAAR
Mb1381|M.bovis_AF2122/97 EHQALRLRMADLQARVDLLRYALHGIAEQGRLELR----TAAAVKVTAAR
MAV_2876|M.avium_104 EHQALRLRMADLQARVDLLQHGLNGIAAQGRLDLR----AAAGVKVTAAR
MSMEG_2130|M.smegmatis_MC2_155 EHQALRMRMADLQARVDMLRYGLAGIAAGGKLDLR----AAAAFKVTAAR
TH_4231|M.thermoresistible__bu EHQALRMRMADLQARTDMLRFALTGIATQGRLDLR----TAAAIKVSAAR
Mvan_1944|M.vanbaalenii_PYR-1 EHQALRIRMADLQARVDLLRHALTGIAAQGRLDLR----AAAGVKVTAAR
MMAR_4532|M.marinum_M SRQSVQNTLAEMARRIDVARVYSRSVVERQLAGEHDLIPQVCFAKNTAVE
MUL_4704|M.ulcerans_Agy99 SRQSVQNTLAEMARRIDVARVYSRSVVERQLAGEHDLIPQVCFAKNTAVE
Mflv_1884|M.gilvum_PYR-GCK ARQTVQSTLAEMARRIDVARVYTRHVVERQLAGDADLITEVCFAKNTAVE
MLBr_00737|M.leprae_Br4923 TFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASD
MAB_1851|M.abscessus_ATCC_1997 KNQHLAYQYAGLAARADMLEVYNRDIAERYMAGEN-VTRKVTIAKLTAGP
* : * : : : . . * *
Rv1346|M.tuberculosis_H37Rv LGEEVISECMHIFGGAGYLVDETTLGKWWRDMKLARVGGGTDEVLWELVA
Mb1381|M.bovis_AF2122/97 LGEEVISECMHIFGGAGYLVDETTLGKWWRDMKLARVGGGTDEVLWELVA
MAV_2876|M.avium_104 LGEEVMSECMHIFGGAGYLVEETPLGRWWRDMKLARVGGGTDEVLWELVA
MSMEG_2130|M.smegmatis_MC2_155 LGEEVISECMHIFGGSGYLVDETPLGKWWRDMKLARVGGGTDEVLWELVA
TH_4231|M.thermoresistible__bu LGEEVLSECMHIFGGSGYLVDETPLGRWWRDMKLARVGGGTDEVLWELVA
Mvan_1944|M.vanbaalenii_PYR-1 LGEEVLSECMHIFGGAGYLVDETPLGRWWRDMKLARVGGGTDEVLWELVA
MMAR_4532|M.marinum_M AGEWVANQAVQLFGGMGYMAES-EIERQYRDMRILGIGGGTTEILTALAA
MUL_4704|M.ulcerans_Agy99 AGEWVANQAVQLFGGMGYMAES-EIERQYRDMRILGIGGGTTEILTALAA
Mflv_1884|M.gilvum_PYR-GCK AGEWVANQAVQLFGGMGYMTES-EVERQYRDMRILGIGGGTTEILTSLAA
MLBr_00737|M.leprae_Br4923 IAMEVTTDAVQLFGGAGYTSDF-PVERFMRDAKITQIYEGTNQIQRVVMS
MAB_1851|M.abscessus_ATCC_1997 LVREAGDWCLQVHGGMGYMTET-WTSRFFRDQRLLSIGGGADEVMMRVLS
. .:::.** ** : : ** :: : *: :: : :
Rv1346|M.tuberculosis_H37Rv AGMTPDHDGYAAVVGASKA--
Mb1381|M.bovis_AF2122/97 AGMTPDHDGYAAVVGASKA--
MAV_2876|M.avium_104 AGMAADHGGYRSVVGASSA--
MSMEG_2130|M.smegmatis_MC2_155 AAMKPDYDGYAEFAGPDAV--
TH_4231|M.thermoresistible__bu AAMKPDYDGYQEIYGADG---
Mvan_1944|M.vanbaalenii_PYR-1 AAMKPDFDSYAEMIGGTTASD
MMAR_4532|M.marinum_M KLLGYQS--------------
MUL_4704|M.ulcerans_Agy99 KLLGYQS--------------
Mflv_1884|M.gilvum_PYR-GCK KTLGYQT--------------
MLBr_00737|M.leprae_Br4923 RALLR----------------
MAB_1851|M.abscessus_ATCC_1997 QIDGFS---------------