For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIWNTPEREELRKTVRSFAEREVLPHIDEWERTGELPRDLHRRAGAAGLLGAGLPESAGGGGGDGADSV IICEQMHQSGAPGGVFASLFTCGIAVPHMIASGDERLIEEFVRPTLAGDKIGALAITEPGGGSDVGHLRT SAVRDGDHFIVNGAKTYITSGVRADFVVTAVRTGGPGAGGVSLLVVEKGTPGFVVSRKLDKMGWRSSDTA ELSYTDARVPAANLVGAENSGFAQIAQAFVSERIALAAQAYSSAQRCLDITVQWCRDRETFGRPLISRQS VQNTLAEMARRIDVARVYSRSVVERQLAGEHDLIPQVCFAKNTAVEAGEWVANQAVQLFGGMGYMAESEI ERQYRDMRILGIGGGTTEILTALAAKLLGYQS
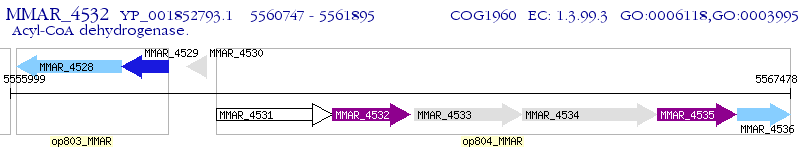
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4532 | fadE13 | - | 100% (382) | acyl-CoA dehydrogenase FadE13 |
| M. marinum M | MMAR_0697 | - | 1e-66 | 38.93% (375) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1989 | fadE20 | 8e-66 | 38.40% (375) | acyl-CoA dehydrogenase FadE20 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1000c | fadE13 | 1e-179 | 86.82% (349) | acyl-CoA dehydrogenase FADE13 |
| M. gilvum PYR-GCK | Mflv_1884 | - | 0.0 | 82.72% (382) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0975c | fadE13 | 0.0 | 87.43% (382) | acyl-CoA dehydrogenase FADE13 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 9e-52 | 32.18% (376) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1073c | - | 0.0 | 80.21% (379) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_1091 | - | 0.0 | 90.84% (382) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5491 | - | 0.0 | 82.20% (382) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4527 | - | 2e-70 | 40.69% (376) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4704 | fadE13 | 0.0 | 98.69% (382) | acyl-CoA dehydrogenase FadE13 |
| M. vanbaalenii PYR-1 | Mvan_4847 | - | 0.0 | 81.35% (386) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4532|M.marinum_M ---------MSIWNTPERE-ELRKTVRSFAEREVLPHIDEWERTGELPRD
MUL_4704|M.ulcerans_Agy99 ---------MSIWNTPERE-ELRKSVRSFAEREVLPHIDEWERTGELPLD
MAV_1091|M.avium_104 ---------MNLWTTPERE-QLRKTVRSFAEREILPHVDEWERSGELPRE
Mb1000c|M.bovis_AF2122/97 ---------MNIWTTPERQ-QLRKTVRAFAEREILPHVDEWERIGELPRG
Rv0975c|M.tuberculosis_H37Rv ---------MNIWTTPERQ-QLRKTVRAFAEREILPHVDEWERIGELPRG
Mflv_1884|M.gilvum_PYR-GCK ---------MSIWTTPERD-DLRRTVRAFAEREVLPFADEWERAREIPRD
Mvan_4847|M.vanbaalenii_PYR-1 ---------MSIWTTPERD-DLRKTVRAFVEREVLPHAGEWERIGELPRE
MSMEG_5491|M.smegmatis_MC2_155 ---------MSIWTTAERE-ALRKTVRAFAEREVLPHAHEWERAGEIPRE
MAB_1073c|M.abscessus_ATCC_199 ------MTAVNSWNTPERK-ALRETVRDFAEREILPNVNEWEREGLLPRE
TH_4527|M.thermoresistible__bu --------MKRTVYEPEHE-AFRETVREYIERELVPHAEKWETERIVDRS
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
.*.. :* ::* *:*: * . : .
MMAR_4532|M.marinum_M LHRRAGAAGLLGAGLPESAGGGGGDGADS-VIICEQMHQSGAPGGVFASL
MUL_4704|M.ulcerans_Agy99 LHRRAGAAGLLGAGLPESAGGGGGDGADS-VIICEQMHQSGAPGGVFASL
MAV_1091|M.avium_104 LHRSAGAAGLLGAGFPESVGGGGGDGADA-VIICEEMHQAGAPGGVFASL
Mb1000c|M.bovis_AF2122/97 LHRLAGAAGLLGAGFPEAVGGGGGDGADP-VIICEEMHQAGAPGGVYASL
Rv0975c|M.tuberculosis_H37Rv LHRLAGAAGLLGAGFPEAVGGGGGDGADP-VIICEEMHQAGAPGGVYASL
Mflv_1884|M.gilvum_PYR-GCK LHRAAAAAGLLGAGFPEEVGGDGGDGADA-VVICEEMHESGCPGGVFASL
Mvan_4847|M.vanbaalenii_PYR-1 LHRKAAEVGLLGAGFPESVGGGGGDGADA-VVICEEMHRAGAPGGVFASL
MSMEG_5491|M.smegmatis_MC2_155 LHRKAAELGLLGAGFPEDAGGSGGDGADP-VVICEEMHYAGSPGGVYASL
MAB_1073c|M.abscessus_ATCC_199 LHRKAGDLGLLGPGSPEAVGGGGGDAIDP-VIVCEELHYAGVPGGVFASL
TH_4527|M.thermoresistible__bu AYVAAGKYGLIGFNMPEKYGGGGTDDFRFNAVIDEEMAKAGVPGPALS--
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAA-CIVIEEVARVDASASLIPAV
*: . ** ** * * :: *:: . .. .
MMAR_4532|M.marinum_M FTCGIAVPHMIASGDERLIEEFVRPTLAGDKIGALAITEPGGGSDVGHLR
MUL_4704|M.ulcerans_Agy99 FTCGIAVPHMIASGDERLIEEFVRPTLAGNKIGALAITEPGGGSDVGHLR
MAV_1091|M.avium_104 FTCGIAVPHMIASGDQRLIEEFVRPTLAGEKIGSLAITEPGGGSDVGHLR
Mb1000c|M.bovis_AF2122/97 FTCGIAVPHMVASGDERLIATYVRPTLAGEKIGALAITEPGGGSDVGHLR
Rv0975c|M.tuberculosis_H37Rv FTCGIAVPHMVASGDERLIATYVRPTLAGEKIGALAITEPGGGSDVGHLR
Mflv_1884|M.gilvum_PYR-GCK FTSGIAVPHMIASQDERLIDTYVRPTLRGENIGSLAITEPGGGSDVGHLT
Mvan_4847|M.vanbaalenii_PYR-1 FTSGIAVPHMIASGDEHLIDTYVRPTLRGEKIGALAITEPGGGSDVGHLT
MSMEG_5491|M.smegmatis_MC2_155 FTCGIAVPHMIASGDQRLIDTYVRPTLRGEKIGALAITEPGGGSDVGHLR
MAB_1073c|M.abscessus_ATCC_199 FTCGISTPHMIASGDQRLIDNYVKPTLAGELIGSLAITEPGGGSDVGHLT
TH_4527|M.thermoresistible__bu LHNDVVAPYFNNLCTEEQKERWMPGIASGETIIAIAMTEPGAGSDLAGIR
MLBr_00737|M.leprae_Br4923 NKLGTMG--LILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMR
. : :. : : : *::* .*** . :
MMAR_4532|M.marinum_M TSAVRDGDHFIVNGAKTYITSGVRADFVVTAVRTG-GPGAGGVSLLVVEK
MUL_4704|M.ulcerans_Agy99 TSAVREGDHFIVNGAKTYITSGVRADFVVTAVRTG-GPGAGGVSLLVVEK
MAV_1091|M.avium_104 TSAVRDGDHYIVNGAKTYITSGVRADYVVTAVRTG-GPGAAGVSLLVVEK
Mb1000c|M.bovis_AF2122/97 TSAVRDGDHYVINGAKTYITSGVRADYVVTAVRTG-GPGAAGVSLLVVEK
Rv0975c|M.tuberculosis_H37Rv TSAVRDGDHYVINGAKTYITSGVRADYVVTAVRTG-GPGAAGVSLLVVEK
Mflv_1884|M.gilvum_PYR-GCK TRAVRDGDCFIVNGAKTYITSGVRADYVVTAARTG-GPGAAGISLIVVDK
Mvan_4847|M.vanbaalenii_PYR-1 TRAVKDGDCYILNGAKTFITSGVRADYVVTAVRTG-GPGAGGISLIVVDK
MSMEG_5491|M.smegmatis_MC2_155 TRADLDGDHYVINGAKTYITSGVRADYVVTAARTG-GPGAGGVSLIVVDK
MAB_1073c|M.abscessus_ATCC_199 TSAKRDGDHYIVNGAKTFITSGVRADYVVTAVRTG-GPGAAGVSLLLIDK
TH_4527|M.thermoresistible__bu TSAVRDGDDWIINGSKTFISSGINCDLVVVVCRTDPEAGHKGFTLFVVER
MLBr_00737|M.leprae_Br4923 TRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHK
* * :** :::** * :*:.* .. .. *. * *.: :::.:
MMAR_4532|M.marinum_M G----TPGFVVSRKLDKMGWRSSDTAELSYTDARVPAANLVGAENSGFAQ
MUL_4704|M.ulcerans_Agy99 G----TPGFVVSRKLDKMGWRSSDTAELSYTDARIPAANLVGAENSGFAQ
MAV_1091|M.avium_104 G----TPGFEVTRKLDKMGWRSSDTAELSYTDVRVPVSNLVGAENTGFAQ
Mb1000c|M.bovis_AF2122/97 D----TPGFEVTRKLDKMGWRSSDTAELCYTDVAVPATNLVGAENSGFTQ
Rv0975c|M.tuberculosis_H37Rv D----TPGFEVTRKLDKMGWRSSDTAELCYTDVAVPATNLVGAENSGFTQ
Mflv_1884|M.gilvum_PYR-GCK G----TPGFAVSRKLDKMGWRSSDTAELSYTDVRVPAANVVGAENTGFLQ
Mvan_4847|M.vanbaalenii_PYR-1 DGPGCSPGLQVTRKLDKMGWRSSDTAELSYTDARVPAGNLVGAENTGFLQ
MSMEG_5491|M.smegmatis_MC2_155 G----TPGFEVTRKLDKMGWRSSDTAELSYTDVRVPVANLVGSENTGFAQ
MAB_1073c|M.abscessus_ATCC_199 D----TPGFEVARKLDKMGWRSSDTAELSFTDVRVPAENLVGSENTGFAQ
TH_4527|M.thermoresistible__bu G----MEGFTRGRKLDKMGLHAQDTAELHFENVRVPHANLLGKEGHGFYH
MLBr_00737|M.leprae_Br4923 D----DEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKT
. *: * .*:* :.. *:** : . :* .::* . **
MMAR_4532|M.marinum_M IAQAFVSERIALAAQAYSSAQRCLDITVQWCRDRETFGRPLISRQSVQNT
MUL_4704|M.ulcerans_Agy99 IAQAFVSERIALAAQAYSSAQRCLDITVQWCRDRETFGRPLISRQSVQNT
MAV_1091|M.avium_104 IAQAFVSERIGLAAQAYSSAQRCLDLTVQWCRDRETFGRPLISRQSVQNT
Mb1000c|M.bovis_AF2122/97 IARAFVSERIGLAAQAYSSAQRCLDLTAQWCRDRETFGRPLISRQSVQNT
Rv0975c|M.tuberculosis_H37Rv IARAFVSERIGLAAQAYSSAQRCLDLTAQWCRDRETFGRPLISRQSVQNT
Mflv_1884|M.gilvum_PYR-GCK IAAAFVSERVGLAAQAYSSAQRCLDLTVEWCRNRQTFGKPLIARQTVQST
Mvan_4847|M.vanbaalenii_PYR-1 IAAAFVSERVGLAAQAYSSAQRCLDLTVEWCRSRETFGKPLITRQSVQNT
MSMEG_5491|M.smegmatis_MC2_155 IAAAFVAERVGLATQAYAGAQRCLDLTVEWCRNRDTFGRPLISRQAVQNT
MAB_1073c|M.abscessus_ATCC_199 IAGAFVSERIGLAAQAYASAQRCLDLTVEWCRNRETFGRPLISRQSVQNT
TH_4527|M.thermoresistible__bu LMQNLPSERLSIAISAIAGARATWRETLQYAKDRKAFGKPIGSFQHNRFL
MLBr_00737|M.leprae_Br4923 ALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFM
: * :. .* . *: : : :.*. **..: : * :
MMAR_4532|M.marinum_M LAEMARRIDVARVYSRSVVERQLAGEHDLIPQVCFAKNTAVEAGEWVANQ
MUL_4704|M.ulcerans_Agy99 LAEMARRIDVARVYSRSVVERQLAGEHDLIPQVCFAKNTAVEAGEWVANQ
MAV_1091|M.avium_104 LAEMARRIDVARVYSRNVVERQLAGETNLIPQVCFAKNTAVEAGEWVANQ
Mb1000c|M.bovis_AF2122/97 LAEMARRIDVARVYAHHVVERQLAGETDLIAQVCFAKNTAVQAGEWVANQ
Rv0975c|M.tuberculosis_H37Rv LAEMARRIDVARVYAHHVVERQLAGETDLIAQVCFAKNTAVQAGEWVANQ
Mflv_1884|M.gilvum_PYR-GCK LAEMARRIDVARVYTRHVVERQLAGDADLITEVCFAKNTAVEAGEWVANQ
Mvan_4847|M.vanbaalenii_PYR-1 LAEMARRIDVARVYTHHVVERELAGETNLIAEVCFAKNTAVDAGEWVAHQ
MSMEG_5491|M.smegmatis_MC2_155 LAGMARRIDVARVYTRHVVERQLAGETNLIAEVCFAKNTAVEAGEWVANQ
MAB_1073c|M.abscessus_ATCC_199 LAEMARRIDVARVYTHALVDRAIAGESNLIAEVCFAKNTAVEAGEWVANQ
TH_4527|M.thermoresistible__bu LAEMDTELEVSEQYIDRCLQAVVDGELTAV-EAAKAKWWCTEVAKKVVDQ
MLBr_00737|M.leprae_Br4923 LADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTD
** * .::.:. *: . :* . : . *. :
MMAR_4532|M.marinum_M AVQLFGGMGYMAESEIERQYRDMRILGIGGGTTEILTALAAKLLGYQS
MUL_4704|M.ulcerans_Agy99 AVQLFGGMGYMAESEIERQYRDMRILGIGGGTTEILTALAAKLLGYQS
MAV_1091|M.avium_104 AVQLFGGMGYMAESEVERQYRDMRILGIGGGTTEILTALSAKLLGFQS
Mb1000c|M.bovis_AF2122/97 AVQLFGGMGYMAESEAN----------ANTGTCESSVSEAAPPK-Y--
Rv0975c|M.tuberculosis_H37Rv AVQLFGGMGYMAESEVERQYRDMRILGIGGGTTEILTALAAKTLGYQS
Mflv_1884|M.gilvum_PYR-GCK AVQLFGGMGYMTESEVERQYRDMRILGIGGGTTEILTSLAAKTLGYQT
Mvan_4847|M.vanbaalenii_PYR-1 AVQLFGGMGYMAESEVERQYRDMRILGIGGGTSEILTALAAKTLGYQS
MSMEG_5491|M.smegmatis_MC2_155 AVQLFGGMGYMAESEVERQYRDMRILGIGGGTTEILTSLAAKTLGFQS
MAB_1073c|M.abscessus_ATCC_199 GVQLFGGMGYMAESEIERQYRDMRIIGIGGGTTEILTSLAAKLLGFQS
TH_4527|M.thermoresistible__bu CVQLHGGYGYMMEYRVARDFVDGRIQTIFGGTTEIMKEIIGRDLGL--
MLBr_00737|M.leprae_Br4923 AVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR--
***.** ** : ** : .