For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTRRALVLAGGGLAGIAWETGVLRGIADESPAAARLLLDSDVLVGTSAGATVAAQISSGCPLDTLYERQ LAETSAEIDPGVDIDAITDLFLTAVTEPHISTRRRLQRIGAVALAVDTVPESVRRQVIAQRLPSHDWPDR VLRVTAIDIATGELVVFHRESNVALVDAVAASCSVPGAWPPVTIAGRRYMDGGVASSVNLGVADDCDAAV VLVPAGADAPSPFGGGAAAEIAAATGMVFAVFADDDSLAAFGPNPLDPLCRVNSAMAGRQQGRREAQAVA RLLGV
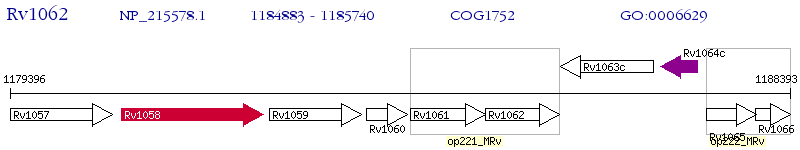
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1062 | - | - | 100% (285) | hypothetical protein Rv1062 |
| M. tuberculosis H37Rv | Rv3239c | - | 7e-06 | 25.00% (216) | transmembrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1091 | - | 1e-159 | 100.00% (285) | hypothetical protein Mb1091 |
| M. gilvum PYR-GCK | Mflv_2029 | - | 1e-102 | 68.68% (281) | patatin |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4758 | - | 4e-11 | 28.97% (214) | patatin-like protein |
| M. marinum M | MMAR_4404 | - | 1e-121 | 77.16% (289) | hypothetical protein MMAR_4404 |
| M. avium 104 | MAV_1189 | - | 1e-117 | 75.09% (289) | phospholipase, patatin family protein |
| M. smegmatis MC2 155 | MSMEG_5285 | - | 1e-100 | 64.56% (285) | phospholipase, patatin family protein |
| M. thermoresistible (build 8) | TH_1491 | - | 6e-87 | 66.67% (252) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1736 | - | 3e-12 | 30.00% (220) | alpha-beta hydrolase superfamily esterase |
| M. vanbaalenii PYR-1 | Mvan_4688 | - | 1e-101 | 66.08% (283) | patatin |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2029|M.gilvum_PYR-GCK ----------------MSGT-RALVLAGGGLAGIAWETGVLLGILEEAPE
Mvan_4688|M.vanbaalenii_PYR-1 ----------------MTDVNRALVLAGGGLAGIAWETGVLLGICDESPD
Rv1062|M.tuberculosis_H37Rv ----------------MTTR-RALVLAGGGLAGIAWETGVLRGIADESPA
Mb1091|M.bovis_AF2122/97 ----------------MTTR-RALVLAGGGLAGIAWETGVLRGIADESPA
MMAR_4404|M.marinum_M ----------------MAPK-RALVLAGGGLAGIAWETGVLLGIADESPA
MAV_1189|M.avium_104 ----------------MTTK-RALVLAGGGIAGIAWETGVLRGIADESPA
MSMEG_5285|M.smegmatis_MC2_155 ----------------MDSK-RALVLAGGGIAGIAWETGILRGIADESPE
TH_1491|M.thermoresistible__bu ----------------VANR-RALVLAGGGLAGIAWETGVLLGIADESPH
MAB_4758|M.abscessus_ATCC_1997 -------------------MTTAFVLSGGASLG-SIQVGMLLALAEAG--
MUL_1736|M.ulcerans_Agy99 MAAVRNSSDMAGNRPYDASVTTAFVLSGGASLG-AIQVGMLQALADEG--
*:**:**. * : :.*:* .: : .
Mflv_2029|M.gilvum_PYR-GCK VGRQLLDSDVLVGTSAGSTVAAQLGSGMGLTELFERQRSESHGAHEIHPG
Mvan_4688|M.vanbaalenii_PYR-1 AGQKLLAADVLLGTSAGSAVAAQVGSGVPLEELFARQLSEEHGAHEIHPG
Rv1062|M.tuberculosis_H37Rv AARLLLDSDVLVGTSAGATVAAQISSGCPLDTLYERQLAETS--AEIDPG
Mb1091|M.bovis_AF2122/97 AARLLLDSDVLVGTSAGATVAAQISSGCPLDTLYERQLAETS--AEIDPG
MMAR_4404|M.marinum_M AARLLLDSDVLVGTSAGSAVAAQISSGTSLDALFDRQIAEQS--AELDPG
MAV_1189|M.avium_104 AARLLTESDVLVGTSAGSAVAAQLCSGHTLDALFDRQVAASS--PEIDSG
MSMEG_5285|M.smegmatis_MC2_155 TAQALLGSDVLVGTSAGSTVAAQLSSGLSLDELYERQVGDAS--AELAPT
TH_1491|M.thermoresistible__bu TADLLLTSDVLVGTSAGSAVAAQLGSGAPLTRLFERQLDPNT--AEIKPR
MAB_4758|M.abscessus_ATCC_1997 -----IAPDLIVGTSVGALNGGWISS--------------------RPDI
MUL_1736|M.ulcerans_Agy99 -----ITPDLIIGTSVGALNGGWIAS--------------------RPDA
.*:::***.*: .. : *
Mflv_2029|M.gilvum_PYR-GCK VSIDAITDLFLKAMTIPDAT----REQKLQKIGAVAAGTDTVGEAVRRKV
Mvan_4688|M.vanbaalenii_PYR-1 VSIDAITDLFLDAMLTQGAT----KAQKLAKIGCVAAETDTVPEAVRRTV
Rv1062|M.tuberculosis_H37Rv VDIDAITDLFLTAVTEPHIS----TRRRLQRIGAVALAVDTVPESVRRQV
Mb1091|M.bovis_AF2122/97 VDIDAITDLFLTAVTEPHIS----TRRRLQRIGAVALAVDTVPESVRRQV
MMAR_4404|M.marinum_M VDIDVITELFLDALREPRIPGVDKIRLRLQRIGSVALATETVSESVRREV
MAV_1189|M.avium_104 VDVEAITELFLTALAEPYDDSEDKTRQQLRRIGAVALATDTVPAPVRRRV
MSMEG_5285|M.smegmatis_MC2_155 AGIGTISELFVDAMLTPGTT----KAQKLRRIGEIAAQTQTVPESVRREV
TH_1491|M.thermoresistible__bu VAIDDLTAEFLEAVRRPGST-----VEKLRRIGAVALSTGTVGEAERRAV
MAB_4758|M.abscessus_ATCC_1997 DGINGLADLWRSLSRKDVFP----TNLSVGFLGFIGQRQSLVSDSGLRRL
MUL_1736|M.ulcerans_Agy99 AGIRALAGLWRSLTRKEVFP----TQPVTGVMGFLGRRQHLVPNTGLRRI
: :: : :* :. * . * :
Mflv_2029|M.gilvum_PYR-GCK IEHRLPSHDWP--ARILRVTAIDIATGELVVLDASSGVTLVDAVAASCAV
Mvan_4688|M.vanbaalenii_PYR-1 IEHRLPAHEWP--QRALRVTAIDIATGELVVFDAASGVGLVDAVAASCAV
Rv1062|M.tuberculosis_H37Rv IAQRLPSHDWP--DRVLRVTAIDIATGELVVFHRESNVALVDAVAASCSV
Mb1091|M.bovis_AF2122/97 IAQRLPSHDWP--DRVLRVTAIDIATGELVVFHRESNVALVDAVAASCSV
MMAR_4404|M.marinum_M IAARLPCHDWP--DRALRVTAIDVSTGELVAFDRDSGVELVDAVAASCAV
MAV_1189|M.avium_104 IAARLPSHRWP--RRALRLTAIDVDTGELTVFDPASGVDLVDAVAASCAV
MSMEG_5285|M.smegmatis_MC2_155 IAARLPSHEWPQ-ERILRIAAIDIATGELVGFDRHSGVDLVDAVAASCAV
TH_1491|M.thermoresistible__bu IAARLPAHDWP--DRDLRLSAVDVGTGELVCFDRDSGVPLVDAVAASCAV
MAB_4758|M.abscessus_ATCC_1997 LKEHLLFRRLEDAPIPLHVVATDVLTGKDVLLS---GGDPIDAIAASAAV
MUL_1736|M.ulcerans_Agy99 LSRELEFTNLEDAPIPLHVVATDVISGTDVLLS---SGNAVDAIAASAAI
: .* *:: * *: :* . : . :**:***.::
Mflv_2029|M.gilvum_PYR-GCK PGVWPPVTIGERRFMDGGVGSTVNMSAAVDCAAAVALVPSPADSPSPWG-
Mvan_4688|M.vanbaalenii_PYR-1 PGVWPPVTIGGRRFMDGGVGSTVNMSAVQDCGAAVVLVPSGANSPSPWG-
Rv1062|M.tuberculosis_H37Rv PGAWPPVTIAGRRYMDGGVASSVNLGVADDCDAAVVLVPAGADAPSPFG-
Mb1091|M.bovis_AF2122/97 PGAWPPVTIAGRRYMDGGVASSVNLGVADDCDAAVVLVPAGADAPSPFG-
MMAR_4404|M.marinum_M PGAWPPVTIAGRRYMDGGVASLVNLGVADDCGVAVVLVPTGADAPSPFG-
MAV_1189|M.avium_104 PGAWPPVAIGGRRYMDGGVASSVNLGVAADCDVAVVLVPSGVDAPSPFG-
MSMEG_5285|M.smegmatis_MC2_155 PGAWPPVTIGDRRYIDGGVSSSINLELAVDCQTAVVFIPAGQSAPSPFDP
TH_1491|M.thermoresistible__bu PGVWPPVTIGERRYMDGGVASTVNMALAADCDTVVALVPSRRDTPSPFG-
MAB_4758|M.abscessus_ATCC_1997 PAVLPPVRIDGRDLMDGGVVNNTPLSHAVALGATEIWVLPTGYACALTET
MUL_1736|M.ulcerans_Agy99 PGIFPPVSINGRELIDGGVVNNTPVSHAVALGADKISVLPTGYSCDLQTL
*. *** * * :**** . : . . : . :
Mflv_2029|M.gilvum_PYR-GCK AGTAAEIEAFPGEALAVFADDDALAAFGPNPLDPACR--APAAEAGRVQG
Mvan_4688|M.vanbaalenii_PYR-1 TGTVAEIEGFPGATLAVFADDESLAAFGANPLDPACR--APAAQAGRAQG
Rv1062|M.tuberculosis_H37Rv GGAAAEIAAATGMVFAVFADDDSLAAFGPNPLDPLCR--VNSAMAGRQQG
Mb1091|M.bovis_AF2122/97 GGAAAEIAAATGMVFAVFADDDSLAAFGPNPLDPLCR--VNSAMAGRQQG
MMAR_4404|M.marinum_M GGSAAEVAEFAGTSLGVFADDDSLAAFGANPLDPRCR--KASALAGRAQG
MAV_1189|M.avium_104 AGPAAEIAAFPGRALAVFADDRSLAAFGPNPLDPGCR--VPSARAGREQG
MSMEG_5285|M.smegmatis_MC2_155 GGIAEEISEFPGAALSIYADDDSLDAFGSNPLDPACR--IPSARAGRAQG
TH_1491|M.thermoresistible__bu PGPAREIDEFGGAAVGVFADDASVAAFGXXXGGHAVRGPVVEALVGLGFS
MAB_4758|M.abscessus_ATCC_1997 PRGAVPMALHAMTLAINQRLVADVNRFEGSVDLRVVPGLCPVRTSPADFS
MUL_1736|M.ulcerans_Agy99 PASAVTLVLHAMTLAVNHRLAVDIERFEQTVDLRVIRPLCPVSISGADFS
. : : * .
Mflv_2029|M.gilvum_PYR-GCK R------REAARVADFLSGL--------------------
Mvan_4688|M.vanbaalenii_PYR-1 R------REARKVAEFLSV---------------------
Rv1062|M.tuberculosis_H37Rv R------REAQAVARLLGV---------------------
Mb1091|M.bovis_AF2122/97 R------REAQAVARLLGV---------------------
MMAR_4404|M.marinum_M R------REASAVAAFLGL---------------------
MAV_1189|M.avium_104 R------REAAAVAEFLGV---------------------
MSMEG_5285|M.smegmatis_MC2_155 R------REAREIAEFLEELAV------------------
TH_1491|M.thermoresistible__bu A------KQAEEATDKVLADDPDASTSTALRSALSMLGRK
MAB_4758|M.abscessus_ATCC_1997 HADELIGRAHEQTRRWLLMPRFKINQADLLEHHHG-----
MUL_1736|M.ulcerans_Agy99 QSATLIPRPHQATREWLATNPARIGQAALLKPHLH-----
: :