For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTAFVLSGGASLGSIQVGMLLALAEAGIAPDLIVGTSVGALNGGWISSRPDIDGINGLADLWRSLSRKD VFPTNLSVGFLGFIGQRQSLVSDSGLRRLLKEHLLFRRLEDAPIPLHVVATDVLTGKDVLLSGGDPIDAI AASAAVPAVLPPVRIDGRDLMDGGVVNNTPLSHAVALGATEIWVLPTGYACALTETPRGAVPMALHAMTL AINQRLVADVNRFEGSVDLRVVPGLCPVRTSPADFSHADELIGRAHEQTRRWLLMPRFKINQADLLEHHH G
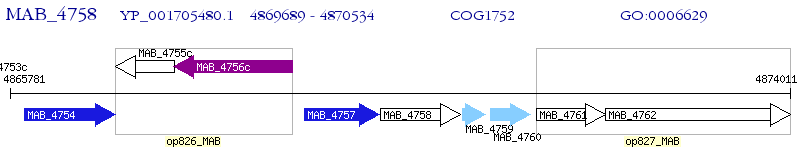
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4758 | - | - | 100% (281) | patatin-like protein |
| M. abscessus ATCC 19977 | MAB_4751 | - | 2e-14 | 34.50% (171) | hypothetical protein MAB_4751 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1092c | - | 3e-13 | 30.00% (180) | hypothetical protein Mb1092c |
| M. gilvum PYR-GCK | Mflv_2030 | - | 1e-14 | 34.25% (181) | patatin |
| M. tuberculosis H37Rv | Rv1063c | - | 3e-13 | 30.00% (180) | hypothetical protein Rv1063c |
| M. leprae Br4923 | MLBr_01423 | - | 3e-07 | 28.57% (196) | hypothetical protein MLBr_01423 |
| M. marinum M | MMAR_2150 | - | 1e-103 | 66.31% (279) | alpha-beta hydrolase superfamily esterase |
| M. avium 104 | MAV_1190 | - | 3e-13 | 30.51% (177) | patatin |
| M. smegmatis MC2 155 | MSMEG_5743 | - | 4e-39 | 36.33% (267) | patatin |
| M. thermoresistible (build 8) | TH_4049 | - | 7e-16 | 50.00% (80) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1736 | - | 1e-101 | 65.59% (279) | alpha-beta hydrolase superfamily esterase |
| M. vanbaalenii PYR-1 | Mvan_4687 | - | 3e-16 | 33.88% (183) | patatin |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2150|M.marinum_M -------------------MTTAFVLSGGASLGAIQVGMLQALADEGITP
MUL_1736|M.ulcerans_Agy99 MAAVRNSSDMAGNRPYDASVTTAFVLSGGASLGAIQVGMLQALADEGITP
MAB_4758|M.abscessus_ATCC_1997 -------------------MTTAFVLSGGASLGSIQVGMLLALAEAGIAP
TH_4049|M.thermoresistible__bu --------------------------------------------------
Mb1092c|M.bovis_AF2122/97 -----MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDI
Rv1063c|M.tuberculosis_H37Rv -----MPAPAALRVRGSSSPRVALALGSGGARGYAHIGVIQALRERGYDI
MAV_1190|M.avium_104 ---------------------MALALGSGGARGYAHIGVIEALQARGYDI
Mflv_2030|M.gilvum_PYR-GCK -------------------MRVALALGSGGARGYAHIGVLNELRERGHEV
Mvan_4687|M.vanbaalenii_PYR-1 -------------------MRVALALGSGGARGYAHIGVLNELTERGHEV
MSMEG_5743|M.smegmatis_MC2_155 ---------------------MAFVLGGGGVLGAAEAGMALALLESGVRP
MLBr_01423|M.leprae_Br4923 ----------MLYLVIVSVVHADLVCQGGGIRGIGLVGAVDALATAGYRF
MMAR_2150|M.marinum_M DLIIGTSVGALNGGWIASRPDA-AGIRALAGLWRSLTRKEVFPTQPVTGL
MUL_1736|M.ulcerans_Agy99 DLIIGTSVGALNGGWIASRPDA-AGIRALAGLWRSLTRKEVFPTQPVTGV
MAB_4758|M.abscessus_ATCC_1997 DLIVGTSVGALNGGWISSRPDI-DGINGLADLWRSLSRKDVFPTNLSVGF
TH_4049|M.thermoresistible__bu --------------------------------------------------
Mb1092c|M.bovis_AF2122/97 VGIAGSSMGAVVG-----------GVHAAGRLDEFAHWAKSLTQRTILRL
Rv1063c|M.tuberculosis_H37Rv VGIAGSSMGAVVG-----------GVHAAGRLDEFAHWAKSLTQRTILRL
MAV_1190|M.avium_104 VGVAGSSMGALVG-----------GLQAAGRLDEFAEWAKSLTQRTILRL
Mflv_2030|M.gilvum_PYR-GCK VGIAGSSMGALVG-----------GLEAAGKTDEFTLWATSLTQRAVLRL
Mvan_4687|M.vanbaalenii_PYR-1 VGIAGSSMGALVG-----------GLQAAGKLDEYTAWASSLTQRAVLRL
MSMEG_5743|M.smegmatis_MC2_155 DLVCGTSVGAINGAALAADPTP-EGARALVKFWDALGGDGVFGGSLIRRA
MLBr_01423|M.leprae_Br4923 PRVAGTSAGALVASLIAALQTAGEPLTRLAEVMRTIDYRKFFDRSLIGRV
MMAR_2150|M.marinum_M MGFLGRRQHLVPN--------TGLRRILSRELEFTNLEDAPIP-------
MUL_1736|M.ulcerans_Agy99 MGFLGRRQHLVPN--------TGLRRILSRELEFTNLEDAPIP-------
MAB_4758|M.abscessus_ATCC_1997 LGFIGQRQSLVSD--------SGLRRLLKEHLLFRRLEDAPIP-------
TH_4049|M.thermoresistible__bu --------------------------------------------------
Mb1092c|M.bovis_AF2122/97 LDPSISAAGILRA--------EKILDAVRDIVGPVAIEQLPIP-------
Rv1063c|M.tuberculosis_H37Rv LDPSISAAGILRA--------EKILDAVRDIVGPVAIEQLPIP-------
MAV_1190|M.avium_104 LDPSISAAGVMRA--------GKILDAVRDILGPVAIEDLRIP-------
Mflv_2030|M.gilvum_PYR-GCK LDPSISSPGVLRA--------EKILESVRDILGEVTIEDLPIP-------
Mvan_4687|M.vanbaalenii_PYR-1 LDPSITSAGVLRA--------EKILDAVRAIVGDVTIEELPIP-------
MSMEG_5743|M.smegmatis_MC2_155 AEVLRRPTSLHDN--------SELRRLLRERLPVHTFEELAVP-------
MLBr_01423|M.leprae_Br4923 PLIGGGLSLLVSDGVYQGAYLEEWLTSLLGDLGVYTFGDLRTGEEPEQFA
MMAR_2150|M.marinum_M --LHVVATDVISGTDVLLSSG-----------NAVDAIAASAAIPGIFPP
MUL_1736|M.ulcerans_Agy99 --LHVVATDVISGTDVLLSSG-----------NAVDAIAASAAIPGIFPP
MAB_4758|M.abscessus_ATCC_1997 --LHVVATDVLTGKDVLLSGG-----------DPIDAIAASAAVPAVLPP
TH_4049|M.thermoresistible__bu --------------------------------------------------
Mb1092c|M.bovis_AF2122/97 --YTAVATDLLAGKSVWFQRG-----------PLDAAIRASIAIPGVIAP
Rv1063c|M.tuberculosis_H37Rv --YTAVATDLLAGKSVWFQRG-----------PLDAAIRASIAIPGVIAP
MAV_1190|M.avium_104 --YTAVATDLLAGKSVWFQRG-----------PLDEAIRASIAIPGVIAP
Mflv_2030|M.gilvum_PYR-GCK --YTSVATDLITGKSVWMQRG-----------RVDDAIRASIAIPGLITP
Mvan_4687|M.vanbaalenii_PYR-1 --YTAVATDLITGKSVWMQRG-----------PLDDAIRASIAIPGVITP
MSMEG_5743|M.smegmatis_MC2_155 --FECVAASIERAREHWFDSG-----------DLVEPVLASCALPGVLPP
MLBr_01423|M.leprae_Br4923 WSLVVTASDLSRRRLVRIPWDLDAYGINPDDFSVARAVHASSAIPYVFEP
MMAR_2150|M.marinum_M VSINGRELIDGGVVNNTPVSHAVALGADKIWVLPTGYSC----DLQTLPA
MUL_1736|M.ulcerans_Agy99 VSINGRELIDGGVVNNTPVSHAVALGADKISVLPTGYSC----DLQTLPA
MAB_4758|M.abscessus_ATCC_1997 VRIDGRDLMDGGVVNNTPLSHAVALGATEIWVLPTGYAC----ALTETPR
TH_4049|M.thermoresistible__bu --------------------------------------------------
Mb1092c|M.bovis_AF2122/97 HEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSE-AGPARDAEP-
Rv1063c|M.tuberculosis_H37Rv HEVDGRLLADGGILDPLPMAPIAGVNADLTIAVSLNGSE-AGPARDAEP-
MAV_1190|M.avium_104 HTLDGRLLADGGILDPLPMAPLSAVNADLTIAVNLSGSE-AITRREAEP-
Mflv_2030|M.gilvum_PYR-GCK HVLGGRLLADGGILDPLPMAPIAAAPADITIAVSLSGEDPAVDRRMQDLR
Mvan_4687|M.vanbaalenii_PYR-1 YVLDGRLLADGGILDPLPMAPIAAVAADMTIAVSISGDDPEASGRGRESR
MSMEG_5743|M.smegmatis_MC2_155 VRIGDEHFFDGGLVNSIPLDRAVSRGADTIWVLHVGRLE----EELRVPR
MLBr_01423|M.leprae_Br4923 VQVAGATWVDGGLLSTFPVELFDRSDGDARWPTFGIRLS----ARPGIPP
MMAR_2150|M.marinum_M SAVTMVLHAMTLAVNHRLAVDIER--FEQTVDLRVIRPLCPVSISG----
MUL_1736|M.ulcerans_Agy99 SAVTLVLHAMTLAVNHRLAVDIER--FEQTVDLRVIRPLCPVSISG----
MAB_4758|M.abscessus_ATCC_1997 GAVPMALHAMTLAINQRLVADVNR--FEGSVDLRVVPGLCPVRTSP----
TH_4049|M.thermoresistible__bu -VLAMALHAFNLAITQRLAVDNTR--FEESIELRGIPQPCPIAISP----
Mb1092c|M.bovis_AF2122/97 NVTAEWLNRMVRSTSALFDVSAARSLLDRPTARAVLSRFGAAAAESDSWS
Rv1063c|M.tuberculosis_H37Rv NVTAEWLNRMVRSTSALFDVSAARSLLDRPTARAVLSRFGAAAAESDSWS
MAV_1190|M.avium_104 AATAEWLTRMMRSTSALFDTAAARSLLDRPTARAVLSRLGGPSGDADAWP
Mflv_2030|M.gilvum_PYR-GCK QPTTDWLSRMWRSSSSLLDTKAARSLLETPTGRAVLGRFVAADDEP----
Mvan_4687|M.vanbaalenii_PYR-1 APSTDWLNRMWRSSSSLLDTKAARSLLDTPTGRAVLGRFAAGDDEP----
MSMEG_5743|M.smegmatis_MC2_155 FLWEVGFAAFEIARRHRFHGDLER--VRSDVAVHVLPSGQP---------
MLBr_01423|M.leprae_Br4923 THPVRGPVSLGIAAIETLVSNQDNAYIDDSCTMLRTIFVPADEVSP----
: : : . .
MMAR_2150|M.marinum_M ----------------------------ADFSQSATLIPRSHQATREWLA
MUL_1736|M.ulcerans_Agy99 ----------------------------ADFSQSATLIPRPHQATREWLA
MAB_4758|M.abscessus_ATCC_1997 ----------------------------ADFSHADELIGRAHEQTRRWLL
TH_4049|M.thermoresistible__bu ----------------------------TDFPHSAELIARSETLARKWLS
Mb1092c|M.bovis_AF2122/97 QAPEIEQRPAGPPADREEAADTPG----LPKMGSFEVMNRTIDIAQSALA
Rv1063c|M.tuberculosis_H37Rv QAPEIEQRPAGPPADREEAADTPG----LPKMGSFEVMNRTIDIAQSALA
MAV_1190|M.avium_104 DNPEEPDQPVLADDD-DEAVELPGAATDVPKLGSFEVMNRTIDIAQAALA
Mflv_2030|M.gilvum_PYR-GCK ----IDAPPEVVKHDSADATGVPE----LPRLGSFEVMNRVIDIAQSALA
Mvan_4687|M.vanbaalenii_PYR-1 ----LPEPDDKVTEEPGNAG--------IPKLGAFEVMNRVIDIAQAALA
MSMEG_5743|M.smegmatis_MC2_155 -----------------------------QRAAATWSNLRYRDSRRIGRR
MLBr_01423|M.leprae_Br4923 ----------------------------IDFNITVEQRQALYQSGLQAGQ
:
MMAR_2150|M.marinum_M TNPARIGQAALLKPHLH---------------------------------
MUL_1736|M.ulcerans_Agy99 TNPARIGQAALLKPHLH---------------------------------
MAB_4758|M.abscessus_ATCC_1997 MPRFKINQADLLEHHHG---------------------------------
TH_4049|M.thermoresistible__bu TPHPATGQAAMLAPHCHGPNRA----------------------------
Mb1092c|M.bovis_AF2122/97 RHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIAVGRALATQALEAFEIDD
Rv1063c|M.tuberculosis_H37Rv RHTLAGYPADLLIEVPRSTCRSLEFHRAVEVIAVGRALATQALEAFEIDD
MAV_1190|M.avium_104 RHTLAVYPPDLLIEVPRSICRGLEFHRAVEVIDAGRALADQALDALEDGD
Mflv_2030|M.gilvum_PYR-GCK RHTLAAYPPDLLIEVPRTVCRSLDFHRAHEVIEIGQELASAALEALDPEP
Mvan_4687|M.vanbaalenii_PYR-1 RHTLAAYPPDLLIEVPRTVCRSLDFHRATEVIEIGQELASAALDTLEQKV
MSMEG_5743|M.smegmatis_MC2_155 AEVAYDATREYLKALP----------------------------------
MLBr_01423|M.leprae_Br4923 QFLKTWNYTEYLTACGRPVTRSAGL-------------------------
*
MMAR_2150|M.marinum_M ----------
MUL_1736|M.ulcerans_Agy99 ----------
MAB_4758|M.abscessus_ATCC_1997 ----------
TH_4049|M.thermoresistible__bu ----------
Mb1092c|M.bovis_AF2122/97 DESAAATIEG
Rv1063c|M.tuberculosis_H37Rv DESAAATIEG
MAV_1190|M.avium_104 DDRPAVER--
Mflv_2030|M.gilvum_PYR-GCK SEA-------
Mvan_4687|M.vanbaalenii_PYR-1 PETASDT---
MSMEG_5743|M.smegmatis_MC2_155 ----------
MLBr_01423|M.leprae_Br4923 ----------