For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNRVSASADDRAAGARPARDLVRVAFGPGVVALGIIAAVTLLQLLIANSDMTGAWGAIASMWLGVHLVPI SIGGRALGVMPLLPVLLMVWATARSTARATSPQSSGLVVRWVVASALGGPLLMAAIALAVIHDASSVVTE LQTPSALRAFTSVLVVHSVGAATGVWSRVGRRALAATALPDWLHDSMRAAAAGVLALLGLSGVVTAGSLV VHWATMQELYGITDSIFGQFSLTVLSVLYAPNVIVGTSAIAVGSSAHIGFATFSSFAVLGGDIPALPILA AAPTPPLGPAWVALLIVGASSGVAVGQQCARRALPFVAAMAKLLVAAVAGALVMAVLGYGGGGRLGNFGD VGVDEGALVLGVLFWFTFVGWVTVVIAGGISRRPKRLRPAPPVELDADESSPPVDMFDGAASEQPPASVA EDVPPSHDDIANGLKAPTADDEALPLSDEPPPRAD
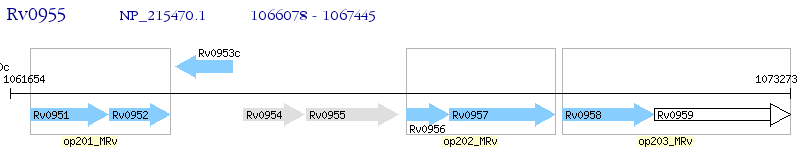
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0955 | - | - | 100% (455) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0980 | - | 0.0 | 100.00% (455) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_1873 | - | 1e-149 | 60.63% (442) | hypothetical protein Mflv_1873 |
| M. leprae Br4923 | MLBr_00159 | - | 1e-176 | 75.89% (419) | hypothetical protein MLBr_00159 |
| M. abscessus ATCC 19977 | MAB_1062 | - | 1e-136 | 56.33% (442) | hypothetical protein MAB_1062 |
| M. marinum M | MMAR_4544 | - | 1e-179 | 73.35% (454) | integral membrane protein |
| M. avium 104 | MAV_1079 | - | 1e-176 | 73.48% (445) | hypothetical protein MAV_1079 |
| M. smegmatis MC2 155 | MSMEG_5517 | - | 1e-154 | 63.46% (457) | hypothetical protein MSMEG_5517 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4714 | - | 1e-178 | 71.90% (459) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_4858 | - | 1e-149 | 62.47% (445) | hypothetical protein Mvan_4858 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1873|M.gilvum_PYR-GCK -------MSTRPVGTRQARDLLRVAFGPSIVALVIIAAIVLLQLLIANSD
Mvan_4858|M.vanbaalenii_PYR-1 -------MSTRPVGTRQARDLLRVAFGPSLVALVIIAAVVLLQLLIANSD
Rv0955|M.tuberculosis_H37Rv MNRVSASADDRAAGARPARDLVRVAFGPGVVALGIIAAVTLLQLLIANSD
Mb0980|M.bovis_AF2122/97 MNRVSASADDRAAGARPARDLVRVAFGPGVVALGIIAAVTLLQLLIANSD
MMAR_4544|M.marinum_M ---MTRVEDSRAAGARQARDLVKVAFAPAVVALVIIAAVTLIQLVIANSD
MUL_4714|M.ulcerans_Agy99 ------MEDSRAAGARQARDLVKVAFAPAVVALVIIAAVTLIQLVIANSD
MLBr_00159|M.leprae_Br4923 ------MGDNRAAGVRQARDLVKVAFGPAVVALAIIAAITLLQLLIANSD
MAV_1079|M.avium_104 ------MEDNRAAGARQARDLVRVAFAPAVVALVIIAAVTLIQLLIANSD
MSMEG_5517|M.smegmatis_MC2_155 -------MNNGPVGTRQARELLRVAFGPSVVALVIIAAVVLLQLVIANSD
MAB_1062|M.abscessus_ATCC_1997 ------MNQPSRANPAQARALLTVAFGPSAVALVIIAAIVLVQLVIANSD
. .. ** *: ***.*. *** ****:.*:**:*****
Mflv_1873|M.gilvum_PYR-GCK MTGALGAIASIWLGVHQVPVSIAGTALGVMPLLPVLLMIYGTARTTAAAT
Mvan_4858|M.vanbaalenii_PYR-1 MTGAFGAIASMWLGVHLVPVSIGGSALGVMPLLPVLVMIYGTARTTAAAT
Rv0955|M.tuberculosis_H37Rv MTGAWGAIASMWLGVHLVPISIGGRALGVMPLLPVLLMVWATARSTARAT
Mb0980|M.bovis_AF2122/97 MTGAWGAIASMWLGVHLVPISIGGRALGVMPLLPVLLMVWATARSTARAT
MMAR_4544|M.marinum_M MTGALGAIASMWLGVHQVPISIGGRELGVMPLFPALLMVWGTARTTALAT
MUL_4714|M.ulcerans_Agy99 MTGALGAIASMWLGVHQVPISIGGRELGVMPLFPAMLMVWGTARTTALAT
MLBr_00159|M.leprae_Br4923 MTGALGAIASMWLGVHQVPIAIGGRELSIMPLLPVLLMVWATAHSTSQAT
MAV_1079|M.avium_104 MTGALGAIASMWLGVHQVPVSIGGRELGVLPLLPVLLMVWATARNTARAT
MSMEG_5517|M.smegmatis_MC2_155 MTGAFGAIASMWLGVHQVPVSIGGRELGVMPLLPVLAMVWGTAKTTAAAT
MAB_1062|M.abscessus_ATCC_1997 MTGTFGAVASMWLGTHLVPISIGGRVIEVLPLLPTAAMVWGVARTVASAL
***: **:**:***.* **::*.* : ::**:*. *::..*:..: *
Mflv_1873|M.gilvum_PYR-GCK TST-SWFVTRWVVASALGGPILFTAICLAVIHDAASVLTELQTPDATRAF
Mvan_4858|M.vanbaalenii_PYR-1 SS--SWFVTRWVVASALGGPVLIAAICLAVIHDAASVLTQLQTPDALHAF
Rv0955|M.tuberculosis_H37Rv SPQSSGLVVRWVVASALGGPLLMAAIALAVIHDASSVVTELQTPSALRAF
Mb0980|M.bovis_AF2122/97 SPQSSGLVVRWVVASALGGPLLMAAIALAVIHDASSVVTELQTPSALRAF
MMAR_4544|M.marinum_M SPYSSWLVVRWVIASALGGPLLIAAICLAVIHDASSVITELQTPSALRAF
MUL_4714|M.ulcerans_Agy99 SPYSSWLVVRWVIASALGGPLLIAAICLAVIHDASSVITELQTPNALRAF
MLBr_00159|M.leprae_Br4923 SAYSSWLVIRWVVASALGGPLLIAAISLAVIHDASSVLTELQTPKALRAF
MAV_1079|M.avium_104 SPHSSWLVIRWVVASALGGPLLMTAVALAVIHDASSVITELQTPSALRAF
MSMEG_5517|M.smegmatis_MC2_155 APNSSWFVTRWVVASALGGPLLIAALLLAVIHDAASVITELQTPSALRAF
MAB_1062|M.abscessus_ATCC_1997 APTTSWYVIRWVIASALAGPLLMTAISLAIIHDASTVLTQLQSPNALRAF
:. * * ***:****.**:*::*: **:****::*:*:**:*.* :**
Mflv_1873|M.gilvum_PYR-GCK GGVFVVHLIGAVLGVGSRLGPRLLALSRLPAWIPDALRGAAAGVIALMGL
Mvan_4858|M.vanbaalenii_PYR-1 GYVFTVHAIGAVLGVGSRVGRRLLSASPLPGWLPDAFRAAAAGVLALMGL
Rv0955|M.tuberculosis_H37Rv TSVLVVHSVGAATGVWSRVGRRALAATALPDWLHDSMRAAAAGVLALLGL
Mb0980|M.bovis_AF2122/97 TSVLVVHSVGAATGVWSRVGRRALAATALPDWLHDSMRAAAAGVLALLGL
MMAR_4544|M.marinum_M VSVLAVHAIGAGLGVWSRVGRRALAVSPLPNWLGDSLRAAAAGVLALLGL
MUL_4714|M.ulcerans_Agy99 VSVLAVHAIGAGLGVWSRVGIRALAVSPLPNWLGDSLRAAAAGVLALLGL
MLBr_00159|M.leprae_Br4923 TGVLVVHAIGAAIGVNSRVGRRVLTASRLPDWVGDSVHAATAGVLALLGL
MAV_1079|M.avium_104 LSVLVVHAIGAGIGVWSQVGPRALAASSLPYWLGDSLRAATAGVLALLGL
MSMEG_5517|M.smegmatis_MC2_155 GGVLAVHAIGALIGVSTKIGRRTLAASPLPKWLPDAVRAASAGVLALFAL
MAB_1062|M.abscessus_ATCC_1997 GCVLGVHAVGAVIGVVTRVGRRIALVLQLPSWPMDAARGAVAGVLALFGL
*: ** :** ** :::* * ** * *: :.* ***:**:.*
Mflv_1873|M.gilvum_PYR-GCK SGAAVAGSLIVHWSTMHDLYSITESMFGQFSLTVLSVLYLPNIVIGAAAV
Mvan_4858|M.vanbaalenii_PYR-1 SGAVVAGSLVVHWSTMHDLFSITDSMFGQLSLTVLSVLYLPNVMVGASAV
Rv0955|M.tuberculosis_H37Rv SGVVTAGSLVVHWATMQELYGITDSIFGQFSLTVLSVLYAPNVIVGTSAI
Mb0980|M.bovis_AF2122/97 SGVVTAGSLVVHWATMQELYGITDSIFGQFSLTVLSVLYAPNVIVGTSAI
MMAR_4544|M.marinum_M SGLVTAGSLVVHWATMQELYGITDSAFGQFSLTVLSVLYVPNVVVGAAAI
MUL_4714|M.ulcerans_Agy99 SGLVTAGSLVVHWATMQELYGITDSAFGQFSLTVLSVLYVPNVVVGAAAI
MLBr_00159|M.leprae_Br4923 SGLVTAGSLVVHWATMQEFYGITDSIFGQFSLTVLSVLYAPNVIVGTSAV
MAV_1079|M.avium_104 SGFVTAGSLVVHWATMQELYGITDSIFGEFSLTVLSMLYAPNVIVGTAAV
MSMEG_5517|M.smegmatis_MC2_155 SGIVTAGSLVVHWGTMHDLFAITDTVFGQLSLTGLSVLYIPNVIVGTSAI
MAB_1062|M.abscessus_ATCC_1997 SGAVTAGSLVVHWATMDQLYSVTNDFVGQLSLTLLAILYAPNVILGSCAL
** ..****:***.**.:::.:*: .*::*** *::** **:::*:.*:
Mflv_1873|M.gilvum_PYR-GCK AVGSSAHIGLATFSSFTVFGGDIPALPVLAAVPTPPLGPVWVALMIIAAV
Mvan_4858|M.vanbaalenii_PYR-1 AVGSSAHIGLATFSSFTVFGGDIPALPVLAAVPTPPLGPVWVALMIIAAV
Rv0955|M.tuberculosis_H37Rv AVGSSAHIGFATFSSFAVLGGDIPALPILAAAPTPPLGPAWVALLIVGAS
Mb0980|M.bovis_AF2122/97 AVGSSAHIGFATFSSFAVLGGDIPALPILAAAPTPPLGPAWVALLIVGAS
MMAR_4544|M.marinum_M AVGASAHIGFATFSAFTVFGGDIPAVPILAAAPTPLLGPVWVALLIIGAS
MUL_4714|M.ulcerans_Agy99 AVGASAHIGFATFSAFTVFGGDIPAVPILAAAPTPPLGPVWVALLIIGAS
MLBr_00159|M.leprae_Br4923 AVGSSAHLGFATFSSFTVFGGDIPALPVLAAAPTPPLAPVWVALLIVGAA
MAV_1079|M.avium_104 AVGSSAHIGFATFSSFTVFGGDIPALPVLAAVPRPPLGPVWVALLIIGAS
MSMEG_5517|M.smegmatis_MC2_155 AVGSSAHIGLATFSSFTVFGGDIPAVPVLAAAPTPPLGPVWVALLIVGAA
MAB_1062|M.abscessus_ATCC_1997 AVGSSAHVGTATFSAFAVFGGQLPAVPILAAVPTPPLGPAWVALMIIGAV
***:***:* ****:*:*:**::**:*:***.* * *.*.****:*:.*
Mflv_1873|M.gilvum_PYR-GCK SAVALGQQCARRPLPLVTALAKVVVAATVAAATMALLGFAGGGALGNFGQ
Mvan_4858|M.vanbaalenii_PYR-1 SAVALGQQCARHPLPMPAALAKVLTAAAAAAGAMALLGHAGGGPLGNFGN
Rv0955|M.tuberculosis_H37Rv SGVAVGQQCARRALPFVAAMAKLLVAAVAGALVMAVLGYGGGGRLGNFGD
Mb0980|M.bovis_AF2122/97 SGVAVGQQCARRALPFVAAMAKLLVAAVAGALVMAVLGYGGGGRLGNFGD
MMAR_4544|M.marinum_M SGVAVGQQCARRPLPPLPAMGKLLAAALVGALAMALLGYGGSGKLGNFGA
MUL_4714|M.ulcerans_Agy99 SGVAVGQQCARRPLPPLPAMGKLLAAALLGALAMALLGYGGSGKLGNFGA
MLBr_00159|M.leprae_Br4923 SGVAVGQQCTRHPLPLLAALAKLLVAAATGALMMALLGYAGSGRLGNFGD
MAV_1079|M.avium_104 SGVAVGQQCARRPLPPVAAMVKLLAASVAGALVMSLLAYAGSGQLGNFGH
MSMEG_5517|M.smegmatis_MC2_155 SAVALGQQCAARPLTPFAALAKLIVASLISAVTMALLGYAGSGRLGNFGH
MAB_1062|M.abscessus_ATCC_1997 SGVAVGQQCARHPVPWPTAIHKVVTAALLAATFLAIVGKLAGGQLGNFGR
*.**:****: :.:. .*: *::.*: .* ::::. ..* *****
Mflv_1873|M.gilvum_PYR-GCK VGVDQSTFAPAVFLWFAGIGGLTVVMSGGVTPRVRKPKPEPVRDADPD--
Mvan_4858|M.vanbaalenii_PYR-1 VGVDQATFAPAVFLWFFGIGALTVAMSGGITPRVRKPKPVPEPAAVVDDA
Rv0955|M.tuberculosis_H37Rv VGVDEGALVLGVLFWFTFVGWVTVVIAGGISRRPKRLRPAPPVELDAD--
Mb0980|M.bovis_AF2122/97 VGVDEGALVLGVLFWFTFVGWVTVVIAGGISRRPKRLRPAPPVELDAD--
MMAR_4544|M.marinum_M VGVDEGALLVGVFFWFAVVGGATLVMAGGIRRRPRRPKPRPAPPVEADVD
MUL_4714|M.ulcerans_Agy99 VGVDEGALLVGVFFWFAVVGGATLVMAGGIRRRPRRPKPRPVPPAEADVD
MLBr_00159|M.leprae_Br4923 IDVDQGALVVGVFFWFAVVGWVTVVVACGIKRFPRHLKPPPALSS-----
MAV_1079|M.avium_104 VGVDQGALLVGVFFWFAVVGGATVVMSGGIRRPTRRLKPRPAPAAPV---
MSMEG_5517|M.smegmatis_MC2_155 VGVDQSTFGPAVFLWFVGIGGLTVVMSGGIVRRVRVAPAAPPPAPEPEPE
MAB_1062|M.abscessus_ATCC_1997 LGIDQGTFGPGVFFWFLVIGLLTVFMLGGATRLPARTRRDPEPAPEPE--
:.:*:.:: .*::** :* *: : * *
Mflv_1873|M.gilvum_PYR-GCK -----------------AADPDAAEFDDQADPDDQAELED--TASGDETP
Mvan_4858|M.vanbaalenii_PYR-1 VIET------------ADAEPHTEEIALEAPPRDRPDPDPRIEPAGEPAP
Rv0955|M.tuberculosis_H37Rv -----------------ESSPPVDMFDGAASEQPPASVAE-DVPPSHDDI
Mb0980|M.bovis_AF2122/97 -----------------ESSPPVDMFDGAASEQPPASVAE-DVPPSHDDI
MMAR_4544|M.marinum_M EPIH-----------EPVDEPFVPLFDQDAPLGDMTSREP-VAPGGDDAD
MUL_4714|M.ulcerans_Agy99 EPIH-----------EPVDEPFVPLFDQDAPLDDVTSREP-VAPGGDDAD
MLBr_00159|M.leprae_Br4923 ------------------EEH-----------ADASSKDH-EAYFG----
MAV_1079|M.avium_104 -----------------AEEHDSVFGEAVGPVGDVAGLDE-IAAEG----
MSMEG_5517|M.smegmatis_MC2_155 PVVPGEVLGEPAEADEPDDEPFGDEPYEETPEEPLVSWHA-DEPPEPAET
MAB_1062|M.abscessus_ATCC_1997 -------------------------MDPEPAPAPEPDPEP-APEPEPQRE
Mflv_1873|M.gilvum_PYR-GCK EPPTEEFAADPPP-----TDRDPEPVPHDAVADTPPVDD-----LDPEE-
Mvan_4858|M.vanbaalenii_PYR-1 PVPDGPIDADLDPEDHFVVDEQTSPFVEDEENGHFVEDEENGPFVEDEEN
Rv0955|M.tuberculosis_H37Rv ANGLKAPTADD------------EALPLSDEPPPRAD-------------
Mb0980|M.bovis_AF2122/97 ANGLKAPTADD------------EALPLSDEPPPRAD-------------
MMAR_4544|M.marinum_M VPPAAPDEADWSDGPDRPDGPDRQDGPDASHPPEHLT-------------
MUL_4714|M.ulcerans_Agy99 VPPAAPDEVDWSDGPD------RQDGPDTSHPPEHLT-------------
MLBr_00159|M.leprae_Br4923 VDLNVPFDLS------------GEDEIPKAEPGEAAD-------------
MAV_1079|M.avium_104 VDEVAAEGVDESPGDA------GEREGDEGDPGERGSGDAGAAGERRPGE
MSMEG_5517|M.smegmatis_MC2_155 DDDAGAPVAPRSPRDLLD-----EDPEDHFFVDEFGDEEVSGDKPRNAED
MAB_1062|M.abscessus_ATCC_1997 PEPESEPEADTDE----------GPEIDPSDPPESAK-------------
Mflv_1873|M.gilvum_PYR-GCK -HFVVDDDMGR-----NGGPPRDH---
Mvan_4858|M.vanbaalenii_PYR-1 GHFVEDEENGLSVVDDDSGGKREQGDH
Rv0955|M.tuberculosis_H37Rv ---------------------------
Mb0980|M.bovis_AF2122/97 ---------------------------
MMAR_4544|M.marinum_M ---------------------------
MUL_4714|M.ulcerans_Agy99 ---------------------------
MLBr_00159|M.leprae_Br4923 ---------------------------
MAV_1079|M.avium_104 ---------------------------
MSMEG_5517|M.smegmatis_MC2_155 ---------------------------
MAB_1062|M.abscessus_ATCC_1997 ---------------------------