For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VALTCTDMSDAVAGSDAEGLTADAIVVGAGLAGLVAACELADRGLRVLILDQENRANVGGQAFWSFGGLF LVNSPEQRRLGIRDSHELALQDWLGTAAFDRPEDYWPEQWAHAYVDFAAGEKRSWLRARGLKIFPLVGWA ERGGYDAQGHGNSVPRFHITWGTGPALVDIFVRQLRDRPTVRFAHRHQVDKLIVEGNAVTGVRGTVLEPS DEPRGAPSSRKSVGKFEFRASAVIVASGGIGGNHELVRKNWPRRMGRIPKQLLSGVPAHVDGRMIGIAQK AGAAVINPDRMWHYTEGITNYDPIWPRHGIRIIPGPSSLWLDAAGKRLPVPLFPGFDTLGTLEYITKSGH DYTWFVLNAKIIEKEFALSGQEQNPDLTGRRLGQLLRSRAHAGPPGPVQAFIDRGVDCVHANSLRELVAA MNELPDVVPLDYETVAAAVTARDREVVNKYSKDGQITAIRAARRYRGDRFGRVVAPHRLTDPKAGPLIAV KLHILTRKTLGGIETDLDARVLKADGTPLAGLYAAGEVAGFGGGGVHGYRALEGTFLGGCIFSGRAAGRG AAEDIR
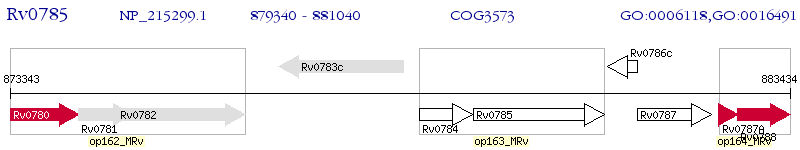
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0785 | - | - | 100% (566) | putative FAD-binding dehydrogenase |
| M. tuberculosis H37Rv | Rv3537 | - | 5e-07 | 40.30% (67) | 3-ketosteroid-delta-1-dehydrogenase |
| M. tuberculosis H37Rv | Rv1817 | - | 2e-06 | 46.88% (64) | hypothetical protein Rv1817 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0808 | - | 0.0 | 99.73% (368) | hypothetical protein Mb0808 |
| M. gilvum PYR-GCK | Mflv_1621 | - | 0.0 | 79.56% (543) | putative FAD-binding dehydrogenase |
| M. leprae Br4923 | MLBr_01226 | nadB | 8e-05 | 34.00% (150) | L-aspartate oxidase |
| M. abscessus ATCC 19977 | MAB_0695 | - | 0.0 | 76.82% (548) | putative FAD-binding dehydrogenase |
| M. marinum M | MMAR_4632 | - | 0.0 | 60.86% (557) | hypothetical protein MMAR_4632 |
| M. avium 104 | MAV_0732 | - | 0.0 | 85.92% (554) | putative FAD-binding dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5835 | - | 0.0 | 81.25% (544) | putative FAD-binding dehydrogenase |
| M. thermoresistible (build 8) | TH_0564 | - | 0.0 | 78.57% (546) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0247 | - | 1e-125 | 51.06% (470) | hypothetical protein MUL_0247 |
| M. vanbaalenii PYR-1 | Mvan_5132 | - | 0.0 | 79.60% (544) | putative FAD-binding dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0785|M.tuberculosis_H37Rv MALTCTDMSDAVAGSDAEGLTADAIVVGAGLAGLVAACELADRGLRVLIL
Mb0808|M.bovis_AF2122/97 --------------------------------------------------
MAV_0732|M.avium_104 -----------MSDSSVAAFGADAIVVGAGLAGLVAACELVDRGLRVLVL
Mflv_1621|M.gilvum_PYR-GCK -------------------MTADVIVVGAGLAGLVAACELAERGRSVLIV
Mvan_5132|M.vanbaalenii_PYR-1 ---------MADVSSAPHVRAADVIVVGAGLAGLVAACELADRGRSVLIV
TH_0564|M.thermoresistible__bu ------------------MADADVIVVGAGLAGLVAACELVDRGRRVLIV
MSMEG_5835|M.smegmatis_MC2_155 ------------------MADADVIVVGAGLAGLVAACELVERGHSVIIV
MAB_0695|M.abscessus_ATCC_1997 ----------------MGDTSTDVIVVGAGLAGLVAACELVDRGKRVVIV
MMAR_4632|M.marinum_M -------------------MDADVIVVGAGLAGLVATHELTRRGKKVAVV
MUL_0247|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01226|M.leprae_Br4923 -----------MMAVPDWKHAADVVVIGTGVAGLAAALAAHRAGRNVVVL
Rv0785|M.tuberculosis_H37Rv DQENRANVGGQAFWSFGGLFLVNSPEQRRLGIRDSHELALQDWLGTAAFD
Mb0808|M.bovis_AF2122/97 --------------------------------------------------
MAV_0732|M.avium_104 DQENSANLGGQAFWSFGGLFFVDSPEQRRLGIRDSHELALQDWLGTAAFD
Mflv_1621|M.gilvum_PYR-GCK DQENSANVGGQAYWSFGGLFFVDSPEQRRLGIRDSHELALQDWLGTAGFD
Mvan_5132|M.vanbaalenii_PYR-1 DQENSANIGGQAYWSFGGLFFVDSPEQRRVGIRDSHELALQDWLGTAGFD
TH_0564|M.thermoresistible__bu DQENAANLGGQAYWSFGGLFFVDSPEQRRLGIRDSYELAWQDWLGTAGFD
MSMEG_5835|M.smegmatis_MC2_155 DQENAANIGGQAFWSFGGLFFVNSPEQRRLGIRDSQELALQDWLGTAGFD
MAB_0695|M.abscessus_ATCC_1997 EQENAANLGGQAYWSFGGLFFVNSPEQRRLGIKDSHELALQDWLGTAGFD
MMAR_4632|M.marinum_M DQENEKNLGGQAFWSFGGLFLVDSPEQRHLGIKDSLELAWNDWSGSAAFD
MUL_0247|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01226|M.leprae_Br4923 TKADQRRGETATHYAQGGIAVVLPGSDDSVDAHASDTLAAGAGMCNLDTV
Rv0785|M.tuberculosis_H37Rv RP--EDYWPEQWAHAYVDFAAGEKRSWLRARGLKIFPLVGWAERGGYDAQ
Mb0808|M.bovis_AF2122/97 --------------------------------------------------
MAV_0732|M.avium_104 RP--EDFWPRQWAHAYVDFAAGEKRSWLRARGLKLFPLVGWAERGGYDAQ
Mflv_1621|M.gilvum_PYR-GCK RA--EDHWPRQWAHAYVDFAAGEKRSWLRDRGLQIFPLVGWAERGGYDAN
Mvan_5132|M.vanbaalenii_PYR-1 RP--EDHWPRQWAHAYVDFAAGEKRSWLRDRGLQIFPLVGWAERGGYGAN
TH_0564|M.thermoresistible__bu RP--EDYWPRKWAEAYVNFATYEKRTWLRARGLQTFPVVGWAERGGYDAR
MSMEG_5835|M.smegmatis_MC2_155 RP--EDHWPREWAHAYVDFAAGEKRSWLRARGLQTFPLVGWAERGGYDAL
MAB_0695|M.abscessus_ATCC_1997 RP--EDHWPRQWAHAYVDFAAGEKRSWLRARGLQTFPVVAWAERGGKDAL
MMAR_4632|M.marinum_M RPDDEDFWATKWARAYVEFAAGEKRSYLTDLGIRFMPTVGWAERGDLRAE
MUL_0247|M.ulcerans_Agy99 ----------------MEFAAGEKRSYLTDLGIRFMPTVGWAERGDLRAE
MLBr_01226|M.leprae_Br4923 YS-----IVAEGYHAVTELVGYGARFDESIPGRWAVTCEGGHSRRRIVHA
Rv0785|M.tuberculosis_H37Rv GHGNSVPRFHITWGTGPALVDIFVRQLRDRP---TVRFAHRHQVDKLIVE
Mb0808|M.bovis_AF2122/97 --------------------------------------------------
MAV_0732|M.avium_104 GHGNSVPRFHITWGTGPALVEIFARRLRDRS---TVRFAHRHRVDELIVE
Mflv_1621|M.gilvum_PYR-GCK GHGNSVPRFHITWGTGPALVDIFARRLIGNP---RVRFAHRHRVDELIVD
Mvan_5132|M.vanbaalenii_PYR-1 GHGNSVPRFHITWGTGPALVDIFARRLIGNP---RVRFAHRHRVDELIVD
TH_0564|M.thermoresistible__bu GHGNSVPRFHITWGTGPAIVDIFARRLHAAAERSQVRFAFRHRVDELIVE
MSMEG_5835|M.smegmatis_MC2_155 GHGNSVPRFHITWGTGPALVEIFARRIRDSV---RVRFAHRHRVDELIVN
MAB_0695|M.abscessus_ATCC_1997 GPGNSVPRFHITWGTGPGLVEIFARRVLAAVDRGQVRIAYRHQVDELVVD
MMAR_4632|M.marinum_M GHGNSVPRFHIAWGTGTGVVEPFVDSALQSAKDKLVTFYHRHRVDELVIT
MUL_0247|M.ulcerans_Agy99 GHGNSVPRFHIAWGTGTGVVEPFVDSALQSAKDKLVTFYHRHRVDELVIT
MLBr_01226|M.leprae_Br4923 GGDATGAEVQRALDHAADVLDIRTSHLALRVLHDGTVVNG---VSVLNPN
Rv0785|M.tuberculosis_H37Rv GNAVTGVRGTVLEPSDEPRGAPSSRKSVGKFEFRASAVIVASGGIGGNHE
Mb0808|M.bovis_AF2122/97 ---MTGVRGTVLEPSDEPRGAPSSRKSVGKFEFRASAVIVASGGIGGNHE
MAV_0732|M.avium_104 GGAVTGVRGSVLEPSAAPRGVASSRNTIAQFEFRADAVIVTSGGIGGNHE
Mflv_1621|M.gilvum_PYR-GCK DGSVIGVRGAVLEPSDAVRGAPSSRNVVGDFEFRGQAVIVASGGIGGNHE
Mvan_5132|M.vanbaalenii_PYR-1 DGAVVGVRGAVLEPSSAVRGAESSRNVVGDFEFRAQAVIVASGGIGGNHE
TH_0564|M.thermoresistible__bu DGVVVGVRGSVLEPSQAPRGFASSRKVVGDFEFRDAAVIVASGGIGGNHD
MSMEG_5835|M.smegmatis_MC2_155 AGLVAGVRGSILEPSNAPRGVASSRKVVGDFEFRASAVIVASGGIGGNLE
MAB_0695|M.abscessus_ATCC_1997 QGAVVGVRGTVLEPSTAARGIASSRIKLGEFEFRAQAVIVTSGGIGGNHD
MMAR_4632|M.marinum_M DGVATGIKGTVLAPDDAERATSSNRDEVGEFELSAQAVIITTGGIGGNHD
MUL_0247|M.ulcerans_Agy99 DGVATGIKGTVLAPDDAERATPSNRDEVGEFELSAQAVIITTGGIGGNHD
MLBr_01226|M.leprae_Br4923 GWGIVSAPSVILASGGLGHLYGATTNPEG--STGDGIALALWAGVAVSDL
. . :* .. : :. . . .: .*:. .
Rv0785|M.tuberculosis_H37Rv LVRKNWPRRMGRIPKQLLSGVPAHVDGRMIGIAQKAGAAVINPDRMWHYT
Mb0808|M.bovis_AF2122/97 LVRKNWPRRMGRIPKQLLSGVPAHVDGRMIGIAQKAGAAVINPDRMWHYT
MAV_0732|M.avium_104 LVRKNWPKRMGRVPEQLLSGVPAHVDGRMIEITQRAGARVINPDRMWHYT
Mflv_1621|M.gilvum_PYR-GCK LVRKYWPERMGRVPEQLLSGVPAHVDGRMLEISATAGASVINSDRMWHYT
Mvan_5132|M.vanbaalenii_PYR-1 LVRKCWPKRMGRVPEQLLSGVPAHVDGRMLEISATAGASVINSDRMWHYT
TH_0564|M.thermoresistible__bu LVRQNWPPRMGRVPEQMLSGVPAYVDGRMMEIAEEAGANVINSDRMWHYT
MSMEG_5835|M.smegmatis_MC2_155 LVRKNWPARLGRVPDQLISGVPAHVDGRMIGIAESAGAHVINNDRMWHYT
MAB_0695|M.abscessus_ATCC_1997 LVRENWPERLGRVPSQLLTGVPAHVDGRMLGISVAAGAHLINRDRMWHYT
MMAR_4632|M.marinum_M MVRRYWPDRMGTPPASMITGVPAYVDGRMLDIANDSGVRLVNRDRMWHYT
MUL_0247|M.ulcerans_Agy99 MVRRYWPDRMGTPPASMITGVPAYVDGRMLDIANDSEVRLVNRDRMWHYT
MLBr_01226|M.leprae_Br4923 EFIQFHPTMLFAAGTGTGGRRPLVTEAIRGEGAILLDSKGNSVTSGVHPL
. . * : * .:. : . *
Rv0785|M.tuberculosis_H37Rv EGITNYDPIWPRHGIRIIP-GPSSLWLDAAGKRLPVPLFPGFDTLGTLEY
Mb0808|M.bovis_AF2122/97 EGITNYDPIWPRHGIRIIP-GPSSLWLDAAGKRLPVPLFPGFDTLGTLEY
MAV_0732|M.avium_104 EGITNYEPIWPRHGIRIIP-GPSSLWLDANGKRLPVPLYPGFDTLGTLEY
Mflv_1621|M.gilvum_PYR-GCK EGITNYDPIWPRHGIRILP-GPSSLWLDATGKRLPVPLYPGFDTLGTLEH
Mvan_5132|M.vanbaalenii_PYR-1 EGITNYDPIWPRHGIRILP-GPSSLWLDAAGKRLPVPLYPGFDTLGTLEY
TH_0564|M.thermoresistible__bu EGITNYDPIWPLHGIRILP-GPSSLWLDAHGKRLPAPLYPGFDTLGTLEY
MSMEG_5835|M.smegmatis_MC2_155 EGITNYDPVWPNHGIRILP-GPSSLWLDANGDRLPVPLYPGYDTLGTLEH
MAB_0695|M.abscessus_ATCC_1997 EGITNHNPIWPGHGIRILP-GPSSLWLDANGSRIPAPLFPGFDTLGTLEH
MMAR_4632|M.marinum_M EGVNNWDPIWPKHAIRIIP-GPSSMWFDALGRRLPAPYLPGYDTLGTLRH
MUL_0247|M.ulcerans_Agy99 EGVNNWDPIWPKHAIRIIP-GPSSMWFDALGRRLPAP-LP--ARLRHLRH
MLBr_01226|M.leprae_Br4923 GDLAPRDVVAAAIDARLKATGDSCVYLDARGIDGFASRFPTVTAACRTVG
.: : : . *: . * *.:::** * .. *
Rv0785|M.tuberculosis_H37Rv ITKSGHD----YTWFVLN--------AKIIEKEFALSGQEQNPDLTGRRL
Mb0808|M.bovis_AF2122/97 ITKSGHD----YTWFVLN--------AKIIEKEFALSGQEQNPDLTGRRL
MAV_0732|M.avium_104 ITKSGFD----YTWFVLN--------AKIIEKEFALSGQEQNPDLTGQSV
Mflv_1621|M.gilvum_PYR-GCK IAQTGQD----YTWFVLN--------SRIIDKEFGLSGQEQNPDLTGRSI
Mvan_5132|M.vanbaalenii_PYR-1 IAQTGQD----YTWFVLN--------SRIIAKEFGLSGQEQNPDLTDRSI
TH_0564|M.thermoresistible__bu ITQTGQD----YTWFILN--------ARIIAKEFALSGQEQNPDLTGRSV
MSMEG_5835|M.smegmatis_MC2_155 ICRSGQD----YTWFILN--------ARIIAKEFALSGQEQNPDLTSRNV
MAB_0695|M.abscessus_ATCC_1997 ICRSGQD----YSWFILN--------ARIIAKEFALSGQEQNPDLTGKDV
MMAR_4632|M.marinum_M LRTTPEIAKYDHSWFILN--------QKIIKKEFALSGSEQNPDITGKSR
MUL_0247|M.ulcerans_Agy99 PAPPAHHRRDRQVRPLLVHLEPEDHQERVRPLGFRAEPRHHRQEPQSRSA
MLBr_01226|M.leprae_Br4923 IDPAREP-------------------IPVVPGAHYSCGGIVTDAYGQTEL
. : .
Rv0785|M.tuberculosis_H37Rv GQLLRS--RAHAGPPGPVQAFIDRGVDCVHANSLRELVAAMNELPDVV-P
Mb0808|M.bovis_AF2122/97 GQLLRS--RAHAGPPGPVQAFIDRGVDFVHANSLRELVAAMNELPDVV-P
MAV_0732|M.avium_104 RELLRN--RARSGPPGPVQAFVDRGVDFVSANSLRELVTAMNKLPDVA-P
Mflv_1621|M.gilvum_PYR-GCK RAVLE---RSRKG-PAPVHAFVDKGVDFVTATSLRELVTRMNAIPGVV-P
Mvan_5132|M.vanbaalenii_PYR-1 RAVLE---RGRKG-PAPVHAFVDKGVDFVTATSLRELVTKMNAVPDVV-P
TH_0564|M.thermoresistible__bu RDVLA---RIRPGAPEPVQAFVDRGADFVSANSLQGLVAAMNELPDVL-P
MSMEG_5835|M.smegmatis_MC2_155 RDLLS---RVKPGAPAPVQAFVDHGVDFVSATSLRDLVAGMNDLPDVV-P
MAB_0695|M.abscessus_ATCC_1997 RGVLR---RGKAGAPAPVQAFVDNGVDFVTATNLPDLVAKMNALPDVS-P
MMAR_4632|M.marinum_M KAVLQERFLNKDATG-PVEAFKRHGADFVVANSLEELVEKMNGLTDEP-L
MUL_0247|M.ulcerans_Agy99 RTVFKQ---GRDRSGRGVQAPRRRLRGRQQPGRTRREDERPDRRTAAGSG
MLBr_01226|M.leprae_Br4923 AGLFAAG---------EVARTGMHGANRLASNSLLEGLVVGGRAGRAAAA
:: * . . . .
Rv0785|M.tuberculosis_H37Rv LDYETVAAAVTARDREVVNKYSKDGQITAIRAARRYRGDRFGRVVAPHRL
Mb0808|M.bovis_AF2122/97 LDYETVAAAVTARDREVVNKYSKDGQITAIRAARRYRGDRFGRVVAPHRL
MAV_0732|M.avium_104 LDYATVEAEVTARDREVANRFSKDGQITAIRAARGYLGDRLGRVVAPHRL
Mflv_1621|M.gilvum_PYR-GCK LEFGVVEAEVTARDREVTNRFTKDGQVTAIRAARAYLPDRISRVVAPHRL
Mvan_5132|M.vanbaalenii_PYR-1 LDFDVVEAEVTARDREVTNRFTKDGQITAIRAARAYLPDRISRVVAPHRL
TH_0564|M.thermoresistible__bu LDYATVAAEVAARDREVVNKFTKDTQVMAIRAARGYLGDRVSRVVAPHRL
MSMEG_5835|M.smegmatis_MC2_155 LDYAKVAAEVTARDREVANRFTKDGQITAIRAARNYLGDRFTRVVAPHRL
MAB_0695|M.abscessus_ATCC_1997 LDYATIAQQVTARDREVANPFGKDPQIAAIRAARAYLADRVARVVAPHRL
MMAR_4632|M.marinum_M LDPAAIRRQIEARDLQVANPFCKDVQVQGIRNARRALGDRLGRVAAPHRI
MUL_0247|M.ulcerans_Agy99 RDPQAD----EARDLQVANPFCKDVQVQGIRNARRALGDRLGRVAAPHRI
MLBr_01226|M.leprae_Br4923 HAAAAGRAYVSKLEPVTHHALKRRELQRVMSRDAAVMRN----AAGLQRL
: . : : : : ... :*:
Rv0785|M.tuberculosis_H37Rv TDPKAGPLIAVKLHILTRKTLGGIETDLDARVLKADGTPLAGLYAAGEVA
Mb0808|M.bovis_AF2122/97 TDPKAGPLIAVKLHILTRKTLGGIETDLDARVLKADGTPLAGLYAAGEVA
MAV_0732|M.avium_104 TDPKAGPLIAVKLHILTRKTLGGIETDLASRVLGADGTPLTGLYAAGEAA
Mflv_1621|M.gilvum_PYR-GCK TDPKAGPLIAVKLHILTRKSLGGLQTDLDSRVLRADGTVLPGLYAAGEAA
Mvan_5132|M.vanbaalenii_PYR-1 TDPKAGPLIAVKLHILTRKSLGGLQTDLDSRVLHDDGTVLPGLYAAGEAA
TH_0564|M.thermoresistible__bu TDSKAGPLIAVKLHILTRKTLGGLETDLDSRVLRGDGEVFDGLYAAGEAA
MSMEG_5835|M.smegmatis_MC2_155 TDPKAGPLIAVKLHILTRKTLGGLETDLDSRVLKEDGTTFGGLYAAGEAA
MAB_0695|M.abscessus_ATCC_1997 TDPKAGPLIAVKLHILTRKTLGGLETDLDSRVLRADGSCFEGLYAAGEVA
MMAR_4632|M.marinum_M LDPKAGPLIATKLHILTRKTLGGIQTDLSSRAIGSNGEPIAGLYAAGEVA
MUL_0247|M.ulcerans_Agy99 LDPKAGPLIATKLHILTRKTLGGIQTDLSSRAIGSNGEPIAGLYAAGEVA
MLBr_01226|M.leprae_Br4923 SDTLAEAPIR---HVTGRRDFEDVALTLTARAVAAAALARNESRGCHHCT
*. * . * *: *: : .: * :*.: . .. . :
Rv0785|M.tuberculosis_H37Rv GFGGGGVHGYRALEGTFLGGCIFSGRAAGRGAAEDIR
Mb0808|M.bovis_AF2122/97 GFGGGGVHGYRALEGTFLGGCIFSGRAAGRGAAEDIR
MAV_0732|M.avium_104 GFGGGGVHGYRALEGTFLGGCIFSGRAAGRGVADDIT
Mflv_1621|M.gilvum_PYR-GCK GFGGGGVHGYRSLEGTFLGGCVFSGRAAGRHAAVAVG
Mvan_5132|M.vanbaalenii_PYR-1 GFGGGGVHGYRSLEGTFLGGCVFSGRAAGRHAASAVG
TH_0564|M.thermoresistible__bu GFGGGGVHGYRSLEGTFLGGCIFSGRAAGRAAARDTA
MSMEG_5835|M.smegmatis_MC2_155 GFGGGGVHGYRSLEGTFLGGCIFSGRAAGRGAAADIA
MAB_0695|M.abscessus_ATCC_1997 GFGGGGVHGYRALEGTFLGGCIFSGRAAGRGAAEDIG
MMAR_4632|M.marinum_M GFGGGGVHGYNALEGTFLGGCIFSGRAAGRAAADAV-
MUL_0247|M.ulcerans_Agy99 GFGGGGVHGYNALEGTFLGGSIFSGRAAGRAAADAV-
MLBr_01226|M.leprae_Br4923 EYPDTAPEHARSTVIRLADDQNLVRAEALATVG----
: . . . .: : .. : * ..