For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSQGDTTSGTQLKPPVEAVRSHYDKSNEFFKLWLDPSMTYSCAYFERPDMTLEEAQYAKRKLALDKLNLE PGMTLLDIGCGWGSTMRHAVAEYDVNVIGLTLSENQYAHDKAMFDEVDSPRRKEVRIQGWEEFDEPVDRI VSLGAFEHFADGAGDAGFERYDTFFKKFYNLTPDDGRMLLHTITIPDKEEAQELGLTSPMSLLRFIKFIL TEIFPGGRLPRISQVDYYSSNAGWKVERYHRIGANYVPTLNAWADALQAHKDEAIALKGQETYDIYMHYL RGCSDLFRDKYTDVCQFTLVK*
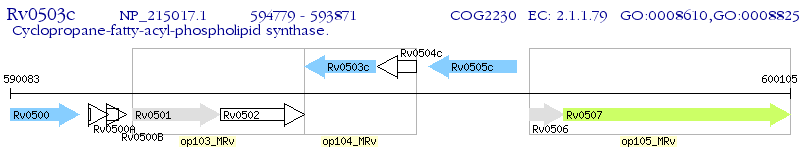
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0503c | cmaA2 | - | 100% (302) | CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 2 CMAA2 (CYCLOPROPANE FATTY ACID SYNTHASE) (CFA SYNTHASE) (CYCLOPROPANE MYCOLIC ACID SYNTHASE 2) (MYCOLIC ACID TRANS-CYCLOPROPANE SYNTHETASE) |
| M. tuberculosis H37Rv | Rv0470c | pcaA | 2e-95 | 58.42% (291) | mycolic acid synthase pcaA (cyclopropane synthase) |
| M. tuberculosis H37Rv | Rv0644c | mmaA2 | 5e-91 | 55.86% (290) | methoxy mycolic acid synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0515c | cmaA2 | 0.0 | 100.00% (302) | cyclopropane-fatty-acyl-phospholipid synthase 2 CMAA2 |
| M. gilvum PYR-GCK | Mflv_3304 | - | 2e-97 | 57.38% (298) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. leprae Br4923 | MLBr_02426 | cmaA2 | 1e-144 | 79.87% (298) | cyclopropane mycolic acid synthase |
| M. abscessus ATCC 19977 | MAB_3881 | - | 1e-89 | 54.79% (292) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. marinum M | MMAR_0831 | cmaA2 | 1e-146 | 80.20% (303) | cyclopropane-fatty-acyl-phospholipid synthase 2 CmaA2 |
| M. avium 104 | MAV_4647 | - | 1e-140 | 78.22% (303) | cyclopropane-fatty-acyl-phospholipid synthase 2 |
| M. smegmatis MC2 155 | MSMEG_1205 | - | 1e-104 | 63.51% (285) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. thermoresistible (build 8) | TH_2761 | - | 1e-101 | 63.73% (284) | PUTATIVE cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. ulcerans Agy99 | MUL_4575 | cmaA2 | 1e-145 | 79.87% (303) | cyclopropane-fatty-acyl-phospholipid synthase 2 CmaA2 |
| M. vanbaalenii PYR-1 | Mvan_3025 | - | 2e-96 | 57.72% (298) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0503c|M.tuberculosis_H37Rv ------MTSQGDT---TSGTQLKPPVEAVRSHYDKSNEFFKLWLDPSMTY
Mb0515c|M.bovis_AF2122/97 ------MTSQGDT---TSGTQLKPPVEAVRSHYDKSNEFFKLWLDPSMTY
MAV_4647|M.avium_104 ------MTSQGE-------TQLKPPVEAVRSHYDKSNEFFKLWLDPSMTY
MMAR_0831|M.marinum_M ------MGKRGDQPSLP--DQLSPPVEAVQSHYDRSNEFFKLFLDPSMTY
MUL_4575|M.ulcerans_Agy99 ------MGKRGDQPSLP--DQLSPPVEAVQSHYDRSNEFFKLFLDPSMTY
MLBr_02426|M.leprae_Br4923 ------MVPSQSHPAKTPRKQLKPPIEAVQSHYDRSNEFFKLWLDPSMTY
MSMEG_1205|M.smegmatis_MC2_155 MKETPQKSNIAVKVGAPPGERHGSTADEVRSHYDLSNEFFRLWQDPTQTY
TH_2761|M.thermoresistible__bu ----------------------------VRSHYDLSNEFFRLWQDPTQTY
Mflv_3304|M.gilvum_PYR-GCK ------MSDTPNGS-----ADLQPHFDDVQSHYDLSDDFYRLFLDPTQTY
Mvan_3025|M.vanbaalenii_PYR-1 ------MPETTSGS-----ADLQPHFEDVQSHYDLSDDFYRLFLDPTQTY
MAB_3881|M.abscessus_ATCC_1997 -------------M-----SKLTPKFSDVQAHYDLSDDFFALFLDPSRTY
*::*** *::*: *: **: **
Rv0503c|M.tuberculosis_H37Rv SCAYFERPDMTLEEAQYAKRKLALDKLNLEPGMTLLDIGCGWGSTMRHAV
Mb0515c|M.bovis_AF2122/97 SCAYFERPDMTLEEAQYAKRKLALDKLNLEPGMTLLDIGCGWGSTMRHAV
MAV_4647|M.avium_104 SCGYWEEGAKTLEEAQYAKRKLALDKLNLEPGMTLLDIGCGWGSTMRHAV
MMAR_0831|M.marinum_M SCAYFERPDMTLEQAQYAKRKLSLDKLNLEPGMTLLDIGCGWGSTMRHAI
MUL_4575|M.ulcerans_Agy99 SCAYFERPDMTLEQAQYAKRKLSLDKLNLEPGMTLLDIGCGWGSTMRHAI
MLBr_02426|M.leprae_Br4923 SCAYFERPDLTLEEAQRAKRDLALSKLGLEPGMTLLDIGCGWGSTMLHAI
MSMEG_1205|M.smegmatis_MC2_155 SCAYFERDDMTLEEAQMAKVDLALGKLGLQPGMTLLDIGCGWGSTIMRAV
TH_2761|M.thermoresistible__bu SCAYFERDDMTLEEAQLAKVDLALGKLGLRPGMTLLDIGCGWGSTIMRAV
Mflv_3304|M.gilvum_PYR-GCK SCAYFERDDMTLEEAQRAKIDLALGKLGLEPGMTLLDIGCGWGGTLLRAL
Mvan_3025|M.vanbaalenii_PYR-1 SCAYFERDDMTLEEAQIAKIDLSLGKLGLQPGMTLLDIGCGWGATLLRAL
MAB_3881|M.abscessus_ATCC_1997 SCAYFEPETLTLEQAQQAKIDLALGKLNLEPGMTLLDVGCGWGSTMKRAL
**.*:* ***:** ** .*:*.**.*.*******:*****.*: :*:
Rv0503c|M.tuberculosis_H37Rv AEYDVNVIGLTLSENQYAHDKAM-FDEVDSP---RRKEVRIQGWEEF-DE
Mb0515c|M.bovis_AF2122/97 AEYDVNVIGLTLSENQYAHDKAM-FDEVDSP---RRKEVRIQGWEEF-DE
MAV_4647|M.avium_104 EEYDVDVIGLTLSENQYAHCVAE-FDKMDSP---RRKEVRIQGWEEFPDE
MMAR_0831|M.marinum_M EEYDVNVIGLTLSENQLVHDERK-FADMDSP---RHKEVRLQGWEQF-DE
MUL_4575|M.ulcerans_Agy99 EEYDVNVIGLTLSENQLVHDERK-FADMDSP---RHKEVRLQSWEQF-DE
MLBr_02426|M.leprae_Br4923 EKYDVNVIGLTLSANQLAHNKLK-FAEIDHTRTDRTKDVRLQGWEQF-DE
MSMEG_1205|M.smegmatis_MC2_155 EKYDVNVIGLTLSHNQRAHIEQR-FAESDSP---RRKEVRLQPWEDF-DE
TH_2761|M.thermoresistible__bu DEHDVDVIGLTLSENQRRHIEEHWFANSDSP---RRKEVRLQGWEDF-DE
Mflv_3304|M.gilvum_PYR-GCK EKYDVNVIGLTLSRNQQAHVQKV-LDEHDSP---RSKTVLLQGWEQF-DQ
Mvan_3025|M.vanbaalenii_PYR-1 DKYDVNVIGLTLSRNQQAHVQKR-LDAHGSG---RSKRVLLAGWEQF-DE
MAB_3881|M.abscessus_ATCC_1997 EHYDVNVIGLTLSKNQFAYVNTL-LDSAGSS---RNYEVRLAGWEEF-EG
.:**:******* ** : : . * * : **:* :
Rv0503c|M.tuberculosis_H37Rv PVDRIVSLGAFEHFADGAGDAGFERYDTFFKKFYNLTPDDGRMLLHTITI
Mb0515c|M.bovis_AF2122/97 PVDRIVSLGAFEHFADGAGDAGFERYDTFFKKFYNLTPDDGRMLLHTITI
MAV_4647|M.avium_104 PIDRIVSLGAFEHFADGAGDAGYERYATFFKKYYDLLPDDGRMLLHSIVV
MMAR_0831|M.marinum_M PVDRIVSLGAFEHFADGAGDASWERYDRFFKMCYNVLPDDGRMLLHTIIV
MUL_4575|M.ulcerans_Agy99 PVDRIVSLGAFEHFADGAGDASWERYDRFFKMCYNVLPDDGRMLLHTIIV
MLBr_02426|M.leprae_Br4923 PVDRIISLGAFEHFADGAGDAGFERYDSFFKMCYDVLPDDGRMLLHTIIV
MSMEG_1205|M.smegmatis_MC2_155 PVDRIVSIGAFEHFG-------FEKYADYFKKTFELMPDDGVMLLHTIVS
TH_2761|M.thermoresistible__bu PVDRIVSIGAFEHFG-------FDNYDFYFKKTYDLLPDDGVMLLHTIVS
Mflv_3304|M.gilvum_PYR-GCK PVDRIVSIGAFEHFG-------SDRYDEFFRRAYSVLPDDGVMLLHTIVK
Mvan_3025|M.vanbaalenii_PYR-1 PVDRIVSIGAFEHFG-------RDRYDEFFKKAYQVLPDGGVMMLHTIIK
MAB_3881|M.abscessus_ATCC_1997 TVDRIVSIGAFEHFG-------HERYDSFFAMAHRVLPSDGKMLLHTIVS
.:***:*:******. :.* :* . : *..* *:**:*
Rv0503c|M.tuberculosis_H37Rv PDKEEAQELGLTSPMSLLRFIKFILTEIFPGGRLPRISQVDYYSSNAGWK
Mb0515c|M.bovis_AF2122/97 PDKEEAQELGLTSPMSLLRFIKFILTEIFPGGRLPRISQVDYYSSNAGWK
MAV_4647|M.avium_104 PTREEGNAMGLKVNMTLLRFISFILKEIYPGGKLPQVNLVDKYATDAGFK
MMAR_0831|M.marinum_M PDAKEAKELGLTAPMSLMRFIKFILTEIFPGGRLPQIPQIDEYSSRAGFK
MUL_4575|M.ulcerans_Agy99 PDAKEAKELGLTAPMSLMRFIKFILTEIFPGGRLPQIPQIDEYSSRAGFK
MLBr_02426|M.leprae_Br4923 PDAKETKELGLTTPMSLLRFIKFILTEIFPGGRLPKISQVDHYSSNAGFT
MSMEG_1205|M.smegmatis_MC2_155 TSKEEVEEMGLPTTMSLMRFFRFIVTEIYPGGRIPLIAMVEDRATEAGFT
TH_2761|M.thermoresistible__bu TSREEVAELGLPTTMSLLRFFKFIVDEIYPGGRIPLIAQVEDAATNAGFR
Mflv_3304|M.gilvum_PYR-GCK PSDEEFAERKLPLTMTKLKFFKFIMDEIFPGGMLPKVDDVEKHATKASFK
Mvan_3025|M.vanbaalenii_PYR-1 PTDEEFADRGLKLTMSIVRFSKFIMDEIFPGGDLPKPTTVAEHATKAGFR
MAB_3881|M.abscessus_ATCC_1997 HHPDEWKRMGIPLLMSRVRFIYFIGTEIFPGGRLPSVPMVDKHAALAGFH
.* : *: ::* ** **:*** :* : :: *.:
Rv0503c|M.tuberculosis_H37Rv VERYHRIGANYVPTLNAWADALQAHKDEAIALKGQETYDIYMHYLRGCSD
Mb0515c|M.bovis_AF2122/97 VERYHRIGANYVPTLNAWADALQAHKDEAIALKGQETYDIYMHYLRGCSD
MAV_4647|M.avium_104 IERHHFIGKNYVPTLTAWGDALEAHKEEAIALKGQETYDIYLKYLRGCAE
MMAR_0831|M.marinum_M VERYHRIGSNYVPTLNSWAQALQANQEKAIELQGQEVYDIYMHYLTGCSD
MUL_4575|M.ulcerans_Agy99 VERYHRIGSNYVPTLNSWAQALQANQEKAIELQGQEVYDIYMHYLTGCSD
MLBr_02426|M.leprae_Br4923 VERYHRIGSHYVPTLNAWAAALEAHKDEAIALQGRQIYDTYMHYLTGCSD
MSMEG_1205|M.smegmatis_MC2_155 VTRKQRLRPHYVRTLDTWAANLEAKKDQAIEITSEEVYERYLKYLKGCAD
TH_2761|M.thermoresistible__bu ITQKQRLRPHYVKTLDAWAANLEANRRAAIEITSEEVYDRYLRYLTGCSD
Mflv_3304|M.gilvum_PYR-GCK VNRIQPLRLHYARTLQIWAASLESRRDDAIAIQSEEVYDRYMKYLTGCAE
Mvan_3025|M.vanbaalenii_PYR-1 LTREQRLREHYAKTLDIWADALRKREDDAVAIQSREDYDRYMKYLTGCAD
MAB_3881|M.abscessus_ATCC_1997 ITRHHSLQPHYARTLDCWATNLAAHEDEAISVQSQEVYDRYIKYLTGCAE
: : : : :*. ** *. * .. *: : ..: *: *::** **::
Rv0503c|M.tuberculosis_H37Rv LFRDKYTDVCQFTLVK---
Mb0515c|M.bovis_AF2122/97 LFRDKYTDVCQFTLVK---
MAV_4647|M.avium_104 LFRDGYTNVCQFTMVK---
MMAR_0831|M.marinum_M LFRDHYTDVCQFTMVK---
MUL_4575|M.ulcerans_Agy99 LFRDHYTDVCQFTMVK---
MLBr_02426|M.leprae_Br4923 LFRDRYTDVCQFTLVK---
MSMEG_1205|M.smegmatis_MC2_155 LFRDGYTDVCQFTCEKSVA
TH_2761|M.thermoresistible__bu LFAKGYTDVCQFTCRKGQ-
Mflv_3304|M.gilvum_PYR-GCK LFTEGYTDVCQFTLAK---
Mvan_3025|M.vanbaalenii_PYR-1 LFRDGYTDICQFTLEK---
MAB_3881|M.abscessus_ATCC_1997 GFREGWIDVVQFTCEK---
* . : :: *** *