For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNDLTPHFEDVQAHYDLSDDFFRLFLDPTQTYSCAHFEREDMTLEEAQIAKIDLALGKLGLQPGMTLLDI GCGWGATMRRAIAQYDVNVVGLTLSKNQAAHVQKSFDEMDTPRDRRVLLAGWEQFNEPVDRIVSIGAFEH FGHDRHADFFARAHKILPPDGVLLLHTITGLTRQQMVDHGLPLTLWLARFLKFIATEIFPGGQPPTIEMV EEQSAKTGFTLTRRQSLQPHYARTLDLWAEALQEHKSEAIAIQSEEVYERYMKYLTGCAKLFRVGYIDVN QFTLAK*
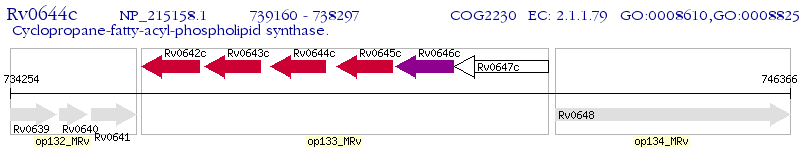
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0644c | mmaA2 | - | 100% (287) | METHOXY MYCOLIC ACID SYNTHASE 2 MMAA2 (METHYL MYCOLIC ACID SYNTHASE 2) (MMA2) (HYDROXY MYCOLIC ACID SYNTHASE) |
| M. tuberculosis H37Rv | Rv3392c | cmaA1 | e-125 | 73.52% (287) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
| M. tuberculosis H37Rv | Rv0470c | pcaA | e-118 | 73.14% (283) | mycolic acid synthase pcaA (cyclopropane synthase) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0663c | mmaA2 | 1e-168 | 99.65% (287) | methoxy mycolic acid synthase |
| M. gilvum PYR-GCK | Mflv_3304 | - | 1e-114 | 67.61% (284) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. leprae Br4923 | MLBr_02459 | umaA2 | 1e-118 | 72.08% (283) | Mycolic acid synthase |
| M. abscessus ATCC 19977 | MAB_3881 | - | 1e-105 | 62.94% (286) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. marinum M | MMAR_3841 | mmaA5 | 1e-141 | 83.97% (287) | methoxy mycolic acid synthase 5 MmaA5 |
| M. avium 104 | MAV_4541 | - | 1e-125 | 74.31% (288) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_1351 | - | 1e-123 | 71.38% (283) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. thermoresistible (build 8) | TH_4640 | - | 1e-126 | 75.62% (283) | PUTATIVE methoxy mycolic acid synthase 5 MmaA5_1 |
| M. ulcerans Agy99 | MUL_3772 | mmaA5 | 1e-141 | 83.62% (287) | methoxy mycolic acid synthase 5 MmaA5 |
| M. vanbaalenii PYR-1 | Mvan_3025 | - | 1e-115 | 68.66% (284) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3304|M.gilvum_PYR-GCK MSDTPNGSADLQPHFDDVQSHYDLSDDFYRLFLDPTQTYSCAYFERD-DM
Mvan_3025|M.vanbaalenii_PYR-1 MPETTSGSADLQPHFEDVQSHYDLSDDFYRLFLDPTQTYSCAYFERD-DM
MLBr_02459|M.leprae_Br4923 ------MSVQLTPHFRNVQAHYDLSDDFFRLFLDPTLTYSCAYFERD-DM
Rv0644c|M.tuberculosis_H37Rv ------MVNDLTPHFEDVQAHYDLSDDFFRLFLDPTQTYSCAHFERE-DM
Mb0663c|M.bovis_AF2122/97 ------MVNDLTPHFEDVQAHYDLSDDFFRLFLDPTQTYSCAHFERE-DM
MMAR_3841|M.marinum_M ------MAKHLAPRFDDVQAHYDLSDDFFRLFLDPTQTYSCAYFERD-DM
MUL_3772|M.ulcerans_Agy99 ------MAKHLAPRFDDVQAHYDLSDDFFRLFLDPTQTYSCAYFERD-DM
TH_4640|M.thermoresistible__bu ---MAEPAKKLKPHFADVQAHYDLSDEFFGLFLDPTRTYSCAYFERD-DM
MAV_4541|M.avium_104 ------MAENLTPHFEEVQAHYDLSDDFFRLFLDPTMTYSCAYWDRNRDI
MSMEG_1351|M.smegmatis_MC2_155 ---MTKQPGKLQPHFDEVQAHYDLSDDFFALFLDPSRTYSCAYFERD-DM
MAB_3881|M.abscessus_ATCC_1997 -------MSKLTPKFSDVQAHYDLSDDFFALFLDPSRTYSCAYFEPE-TL
.* *:* :**:******:*: *****: *****::: : :
Mflv_3304|M.gilvum_PYR-GCK TLEEAQRAKIDLALGKLGLEPGMTLLDIGCGWGGTLLRALEKYDVNVIGL
Mvan_3025|M.vanbaalenii_PYR-1 TLEEAQIAKIDLSLGKLGLQPGMTLLDIGCGWGATLLRALDKYDVNVIGL
MLBr_02459|M.leprae_Br4923 TLEEAQLAKVDLALGKLKLEPGMTLLDIGCGWGATMKRAIEIYDVNVIGL
Rv0644c|M.tuberculosis_H37Rv TLEEAQIAKIDLALGKLGLQPGMTLLDIGCGWGATMRRAIAQYDVNVVGL
Mb0663c|M.bovis_AF2122/97 TLEEAQIAKIDLALGKLGLQPGMTLLDIGCGWGATMRRAIAQYDVNVVGL
MMAR_3841|M.marinum_M TLEEAQIAKIDLALGKLGLEPGMTLLDIGCGWGATMRRAIEKYDVNVVGL
MUL_3772|M.ulcerans_Agy99 TLEEAQIAKIDLALGKLGLEPGMTLLDIGCGWGATMRRAIEKYDVNVVGL
TH_4640|M.thermoresistible__bu SLEEAQLAKIDLALGKLGLQPGMTLLDIGCGWGSTMLRALERHDVDVVGL
MAV_4541|M.avium_104 SLEEAQLRKIDLSLGKLGLRPGMTLLDVGCGWGATMRRAVENYDVNVIGL
MSMEG_1351|M.smegmatis_MC2_155 TLEEAQLAKIDLSLGKLGLQPGMTLLDVGCGWGATMLRALERYDVNVVGL
MAB_3881|M.abscessus_ATCC_1997 TLEQAQQAKIDLALGKLNLEPGMTLLDVGCGWGSTMKRALEHYDVNVIGL
:**:** *:**:**** *.*******:*****.*: **: :**:*:**
Mflv_3304|M.gilvum_PYR-GCK TLSRNQQAHVQKVLDEHDSPRSKTVLLQGWEQFDQPVDRIVSIGAFEHFG
Mvan_3025|M.vanbaalenii_PYR-1 TLSRNQQAHVQKRLDAHGSGRSKRVLLAGWEQFDEPVDRIVSIGAFEHFG
MLBr_02459|M.leprae_Br4923 TLSENQAAHVQKTFDELDTPRTRRVLLEGWEKFHEPVDRIVSIGAFEHFG
Rv0644c|M.tuberculosis_H37Rv TLSKNQAAHVQKSFDEMDTPRDRRVLLAGWEQFNEPVDRIVSIGAFEHFG
Mb0663c|M.bovis_AF2122/97 TLSKNQAAHVQKSFDEMDTPLDRRVLLAGWEQFNEPVDRIVSIGAFEHFG
MMAR_3841|M.marinum_M TLSKNQAAHVQKSFDQLDTARTRRVLLEGWEQFDEPVDRIVSIGAFEHFG
MUL_3772|M.ulcerans_Agy99 TLSKNQAAHVQKSFDQLDTARTRRVLLEGWEQFDEPVDRIVSIGAFEHFG
TH_4640|M.thermoresistible__bu TLSANQHAHVEQLLADSPSRRSRRVLLRGWEEFDEPVDRIVSIGAFEHFG
MAV_4541|M.avium_104 TLSRHQAAHVQQLLDAMDTPRSRRVLLHGWEEFDEPVDRIVSIGAFEHFG
MSMEG_1351|M.smegmatis_MC2_155 TLSRNQQAHVERRFSESSSPRTKRVLLAGWEEFDEPVDRIVSIGAFEHFG
MAB_3881|M.abscessus_ATCC_1997 TLSKNQFAYVNTLLDSAGSSRNYEVRLAGWEEFEGTVDRIVSIGAFEHFG
*** :* *:*: : : * * ***:*. .**************
Mflv_3304|M.gilvum_PYR-GCK SDRYDEFFRRAYSVLPDDGVMLLHTIVKPSDEEFAERKLPLTMTKLKFFK
Mvan_3025|M.vanbaalenii_PYR-1 RDRYDEFFKKAYQVLPDGGVMMLHTIIKPTDEEFADRGLKLTMSIVRFSK
MLBr_02459|M.leprae_Br4923 RQRYARFFKMAHEVLPADSVMLLHTIVRPTFKDARARGMTLTHEIVHFTQ
Rv0644c|M.tuberculosis_H37Rv HDRHADFFARAHKILPPDGVLLLHTITGLTRQQMVDHGLPLTLWLARFLK
Mb0663c|M.bovis_AF2122/97 HDRHADFFARAHKILPPDGVLLLHTITGLTRQQMVDHGLPLTLWLARFLK
MMAR_3841|M.marinum_M HDRYDDFFTLAHNILPSDGVMLLHTITGLTMPQMVENGLPLTLWLARFLK
MUL_3772|M.ulcerans_Agy99 HDRYDDFFTLAHNILPSDGVMLLHTITGLTMPQMVENGLPLTLWLARFLK
TH_4640|M.thermoresistible__bu HDRYDDFFATAHRLLPDDGVMLLHTITALTVPQMVDRGIPLTFEVARFVK
MAV_4541|M.avium_104 YDRYDDFFATAYRVLPDDGVMLLHTITMLTPEQIVERGLPLTEEQTSFNE
MSMEG_1351|M.smegmatis_MC2_155 HERYDDFFRFAHRVLPDDGVMLLHTITGIHPKHAQELGVPLTFEMARFIR
MAB_3881|M.abscessus_ATCC_1997 HERYDSFFAMAHRVLPSDGKMLLHTIVSHHPDEWKRMGIPLLMSRVRFIY
:*: ** *: :** .. ::**** . : * *
Mflv_3304|M.gilvum_PYR-GCK FIMDEIFPGGMLPKVDDVEKHATKASFKVNRIQPLRLHYARTLQIWAASL
Mvan_3025|M.vanbaalenii_PYR-1 FIMDEIFPGGDLPKPTTVAEHATKAGFRLTREQRLREHYAKTLDIWADAL
MLBr_02459|M.leprae_Br4923 FILAEIFPGGWLPTVPTVEEHATAAGFKFTRIQSLQLHYAKTLDLWAARL
Rv0644c|M.tuberculosis_H37Rv FIATEIFPGGQPPTIEMVEEQSAKTGFTLTRRQSLQPHYARTLDLWAEAL
Mb0663c|M.bovis_AF2122/97 FIATEIFPGGQPPTIEMVEEQSAKTGFTLTRRQSLQPHYARTLDLWAEAL
MMAR_3841|M.marinum_M FIQTEIFPGGHPPTIEMVEEQSVKPGFTLTRRHSLQLHYAKTLDLWAEAL
MUL_3772|M.ulcerans_Agy99 FIQTEIFPGGHPPTIEMVEEQSVKPGFTLTRRHSLQLHYAKTLDLWAEAL
TH_4640|M.thermoresistible__bu FILTEIFPGGRLPSIEKVEDHSGANGFTLTRRQSLQSHYARTLDLWAEAL
MAV_4541|M.avium_104 FIATEIFPGGQLPAIEVVEFHSSKMGFTLKRRQSLQLHYARTLDHWADAL
MSMEG_1351|M.smegmatis_MC2_155 FIVTEIFPGGRLPTVDMVIDHATRASFTLERRQSLQLHYARTLDMWAAAL
MAB_3881|M.abscessus_ATCC_1997 FIGTEIFPGGRLPSVPMVDKHAALAGFHITRHHSLQPHYARTLDCWATNL
** ****** * * :: .* . * : *: ***:**: ** *
Mflv_3304|M.gilvum_PYR-GCK ESRRDDAIAIQSEEVYDRYMKYLTGCAELFTEGYTDVCQFTLAK------
Mvan_3025|M.vanbaalenii_PYR-1 RKREDDAVAIQSREDYDRYMKYLTGCADLFRDGYTDICQFTLEK------
MLBr_02459|M.leprae_Br4923 EANKNKAIAIQSEEVYDRYMKYLTGCAKLFRDGYTDVDQFTLEKQPATAR
Rv0644c|M.tuberculosis_H37Rv QEHKSEAIAIQSEEVYERYMKYLTGCAKLFRVGYIDVNQFTLAK------
Mb0663c|M.bovis_AF2122/97 QEHKSEAIAIQSEEVYERYMKYLTGCAKLFRVGYIDVNQFTLAK------
MMAR_3841|M.marinum_M EARKDEAIKIQSDEVYERYMRYLTGCSKLFQEGYIDVNQFTLQKAR----
MUL_3772|M.ulcerans_Agy99 EVRKDEAIKIQSDEVYERYMRYLTGCSKLFQEGYVDVNQFTLQKAR----
TH_4640|M.thermoresistible__bu QQHREEAITVQSDEVYDRYIKYLTGCAKGFRVGYIDVNQFTLQK------
MAV_4541|M.avium_104 RAHHSEAVAIQSEEVYQRYMRYLTGCADAFRAGYIDVNQFTLAK------
MSMEG_1351|M.smegmatis_MC2_155 ESHHDDAVRIQSQEVYDRYMHYLTGCANAFRVGYIDVNQFTLAK------
MAB_3881|M.abscessus_ATCC_1997 AAHEDEAISVQSQEVYDRYIKYLTGCAEGFREGWIDVVQFTCEK------
....*: :** * *:**::*****:. * *: *: *** *
Mflv_3304|M.gilvum_PYR-GCK --
Mvan_3025|M.vanbaalenii_PYR-1 --
MLBr_02459|M.leprae_Br4923 RH
Rv0644c|M.tuberculosis_H37Rv --
Mb0663c|M.bovis_AF2122/97 --
MMAR_3841|M.marinum_M --
MUL_3772|M.ulcerans_Agy99 --
TH_4640|M.thermoresistible__bu --
MAV_4541|M.avium_104 --
MSMEG_1351|M.smegmatis_MC2_155 --
MAB_3881|M.abscessus_ATCC_1997 --