For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PPLDITDERLTREDTGYHKGLHSRQLQMIALGGAIGTGLFLGAGGRLASAGPGLFLVYGICGIFVFLILR ALGELVLHRPSSGSFVSYAREFYGEKVAFVAGWMYFLNWAMTGIVDTTAIAHYCHYWRAFQPIPQWTLAL IALLVVLSMNLISVRLFGELEFWASLIKVIALVTFLIVGTVFLAGRYKIDGQETGVSLWSSHGGIVPTGL LPIVLVTSGVVFAYAAIELVGIAAGETAEPAKIMPRAINSVVLRIACFYVGSTVLLALLLPYTAYKEHVS PFVTFFSKIGIDAAGSVMNLVVLTAALSSLNAGLYSTGRILRSMAINGSGPRFTAPMSKTGVPYGGILLT AGIGLLGIILNAIKPSQAFEIVLHIAATGVIAAWATIVACQLRLHRMANAGQLQRPKFRMPLSPFSGYLT LAFLAGVLILMYFDEQHGPWMIAATVIGVPALIGGWYLVRNRVTAVAHHAIDHTKSVAVVHSADPI*
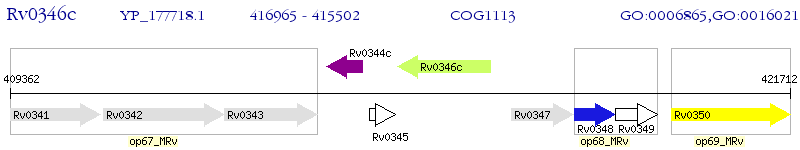
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0346c | ansP2 | - | 100% (487) | POSSIBLE L-ASPARAGINE PERMEASE ANSP2 (L-ASPARAGINE TRANSPORT PROTEIN) |
| M. tuberculosis H37Rv | Rv2127 | ansP1 | 0.0 | 73.16% (462) | L-asparagine permease ansP1 |
| M. tuberculosis H37Rv | Rv1704c | cycA | 1e-81 | 39.91% (431) | D-serine/alanine/glycine transporter protein CycA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0354c | ansP2 | 0.0 | 100.00% (487) | L-asparagine ABC transporter permease |
| M. gilvum PYR-GCK | Mflv_1998 | - | 6e-75 | 36.30% (416) | amino acid permease-associated region |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 0.0 | 71.00% (462) | putative L-asparagine permease |
| M. abscessus ATCC 19977 | MAB_2914 | - | 1e-179 | 64.32% (468) | L-asparagine permease 1 (L-asparagine transport) |
| M. marinum M | MMAR_0627 | ansP1_1 | 0.0 | 83.51% (473) | L-asparagine permease AnsP1_1 |
| M. avium 104 | MAV_4626 | - | 6e-74 | 36.54% (416) | gaba permease |
| M. smegmatis MC2 155 | MSMEG_5473 | - | 4e-73 | 35.02% (414) | transporter |
| M. thermoresistible (build 8) | TH_0471 | - | 1e-18 | 24.72% (441) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_0383 | ansP1 | 0.0 | 67.01% (485) | L-asparagine permease AnsP1 |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 4e-18 | 24.37% (439) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1998|M.gilvum_PYR-GCK -------------------------MSAAPTLKKALNQRQLTMIAIGGVI
MSMEG_5473|M.smegmatis_MC2_155 -------------------------MGDTPALKRALSQRQLRMIAIGGVI
MAV_4626|M.avium_104 -------------------------MAGTSQLRQGLSQRQLSMIALGGVI
Rv0346c|M.tuberculosis_H37Rv ------MPPLDITD--------ERLTREDTGYHKGLHSRQLQMIALGGAI
Mb0354c|M.bovis_AF2122/97 ------MPPLDITD--------ERLTREDTGYHKGLHSRQLQMIALGGAI
MMAR_0627|M.marinum_M ------MPPLDTAAT-------ARLTREDRGYHKSLKNRQLQMIALGGAI
MLBr_01305|M.leprae_Br4923 ------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAI
MUL_0383|M.ulcerans_Agy99 ------MRTPSSPTHP----DDIALASEDAGYHKSLKPRQLQMIAIGGAI
MAB_2914|M.abscessus_ATCC_1997 ------MTSPATYDT-------SAPAPEEQGYHKGLNNRQVQMIGIGGAI
TH_0471|M.thermoresistible__bu --------MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVVI
Mvan_1740|M.vanbaalenii_PYR-1 MVQEPGAQTVGRRRTKSVEQSIADTDEPSTRLRKELNWWDLTVFGVSVVI
:: * :: ::.:. .*
Mflv_1998|M.gilvum_PYR-GCK GAGLFVGSGVVIA-DTGPGAFLTYGMAGVLIIMVMRMLAEMAVANPSTGS
MSMEG_5473|M.smegmatis_MC2_155 GAGLFVGSGVVIG-DTGPGTFITYGLAGVLIIMVMRMLAEMAVANPSTGS
MAV_4626|M.avium_104 GAGLFVGSGVVIK-DTGPAAFLTYALCGLLIVLVMRMLGEMAAANPSTGS
Rv0346c|M.tuberculosis_H37Rv GTGLFLGAGGRLA-SAGPGLFLVYGICGIFVFLILRALGELVLHRPSSGS
Mb0354c|M.bovis_AF2122/97 GTGLFLGAGGRLA-SAGPGLFLVYGICGIFVFLILRALGELVLHRPSSGS
MMAR_0627|M.marinum_M GTGLFLGAGGRLA-SAGPGLFLVYGICGFFVFLILRALGELVLHRPSSGS
MLBr_01305|M.leprae_Br4923 GTGLFLGAGGRLA-KAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGS
MUL_0383|M.ulcerans_Agy99 GTGLFLGAGGRLA-SAGPALFLVYAVCGVFAFLILRALSELVLHRPSSGS
MAB_2914|M.abscessus_ATCC_1997 GTGLFMGAGGRLH-SAGPGLFLVYAVCGVFVFFILRALGELILHRPSSGS
TH_0471|M.thermoresistible__bu GAGIFTVTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGS
Mvan_1740|M.vanbaalenii_PYR-1 GAGIFTITASTAANITGPAISLSFIIAAIACGLAALCYAEFASTVPVAGS
*:*:* :. :**. : : :... : .*: * :**
Mflv_1998|M.gilvum_PYR-GCK FADYSRNALGHWAGFSVGWLYWYFWVIVVGFEAIAGAKIVQYW---MDVP
MSMEG_5473|M.smegmatis_MC2_155 FADYSRNALGNWAGFSVGWLYWYFWVIVVGFEAIAGAKIIQFW---IDVP
MAV_4626|M.avium_104 FADYAAAALGRWAGFSVAWLYWYFWVIVVGFEAVAGGKVLNYW---FHAP
Rv0346c|M.tuberculosis_H37Rv FVSYAREFYGEKVAFVAGWMYFLNWAMTGIVDTTAIAHYCHYWRAFQPIP
Mb0354c|M.bovis_AF2122/97 FVSYAREFYGEKVAFVAGWMYFLNWAMTGIVDTTAIAHYCHYWRAFQPIP
MMAR_0627|M.marinum_M FVSYAREFFGEKVAFVAGWMYFLNWAMTGVVDTTAIAHYCHYWKALQFVP
MLBr_01305|M.leprae_Br4923 FVSYAREFFGEKAAYVVGWLYFLDWAMTAIVDTTAIATYLHRWTIFTALP
MUL_0383|M.ulcerans_Agy99 FVSYAREFFGEKAAYVAGWMYFLNWAMTSIVDSTAIATYFHFWRVFEGIP
MAB_2914|M.abscessus_ATCC_1997 FVSYAREFLGEKAAYVAGWMYFFNWAATAIVDVTAIALYMHYWSAFKAIP
TH_0471|M.thermoresistible__bu AYTFSYATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLGEVFEFAGAT
Mvan_1740|M.vanbaalenii_PYR-1 AYTFSYATFGEFVAWIIGWDLILEFAVAAAVVAKGWSSYLGTVFGFAGGT
:: *. ..: .* : . . . . .
Mflv_1998|M.gilvum_PYR-GCK LWLS-------ALIFMVLMTATNLFSVASYGEFEFWFAGIKVAAIIAFIA
MSMEG_5473|M.smegmatis_MC2_155 LWLT-------ALILLIAMTATNLFSVSSFGEFEYWFAGVKVAAILAFIG
MAV_4626|M.avium_104 LWVL-------SLGLMLLMTATNLFSVSSFGEFEFWFAGIKVATIVIFLV
Rv0346c|M.tuberculosis_H37Rv QWTL-------ALIALLVVLSMNLISVRLFGELEFWASLIKVIALVTFLI
Mb0354c|M.bovis_AF2122/97 QWTL-------ALIALLVVLSMNLISVRLFGELEFWASLIKVIALVTFLI
MMAR_0627|M.marinum_M QWTL-------ALVALVSVLLMNLISVKLFGELEFWAALIKVIALVIFLI
MLBr_01305|M.leprae_Br4923 QWTL-------ALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAFLV
MUL_0383|M.ulcerans_Agy99 QWAL-------ALFALGIALSMNLISETLFGEIEFWAALIKVVSLLTFRV
MAB_2914|M.abscessus_ATCC_1997 QWSI-------ALIALVIVLTMNTISVKLFGEMEFWAALIKVVAIMGFLV
TH_0471|M.thermoresistible__bu TRLGPITLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVI
Mvan_1740|M.vanbaalenii_PYR-1 TEVAGFQLDWGALVIIALVTLILVMGTKLSAGVSLAITAVKVSVVLLVVI
:* : . . .. : :** :: .
Mflv_1998|M.gilvum_PYR-GCK VGAAFVLG---------LWPGKGADFSNLTSHGGFMP------LGFGAIT
MSMEG_5473|M.smegmatis_MC2_155 LGAFYVLG---------VWPDKDLDFSNLTAHGGFFP------LGATAVT
MAV_4626|M.avium_104 VGAAYVIG---------LVPGHHDGLTNLVSHGGFFP------RGVGAVF
Rv0346c|M.tuberculosis_H37Rv VGTVFLAGR-------YKIDGQETGVSLWSSHGGIVP------TGLLPIV
Mb0354c|M.bovis_AF2122/97 VGTVFLAGR-------YKIDGQETGVSLWSSHGGIVP------TGLLPIV
MMAR_0627|M.marinum_M IGSIFLAGR-------YHVDGQETGLGLWRSHGGFLP------TGLVPLV
MLBr_01305|M.leprae_Br4923 VGTIFLGGR-------YPVDGHNTGLSLWTSHGGLFP------TGVAPLI
MUL_0383|M.ulcerans_Agy99 VGTIFLAGR-------YPVDGHSTGLGLLAHGGGLFP------YGLLPLV
MAB_2914|M.abscessus_ATCC_1997 IGTVFLAGR-------FTVEGASTGPSVIADNGGLLP------TGLMALV
TH_0471|M.thermoresistible__bu VGAFFIKAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYGLL
Mvan_1740|M.vanbaalenii_PYR-1 VGAFYIKAENFSPFVPAPEEGGSGGTGTEQSLFSLLTGAEGSHYGWYGVL
:*: :: . . . .:.. * :
Mflv_1998|M.gilvum_PYR-GCK VGIVTVIFSMVGAEIATIAAAESQDPERAVAKAANSVILRILIFYVGAVL
MSMEG_5473|M.smegmatis_MC2_155 VGVVTVIFSMVGAEIATIAAAESSDPERAVAKAANSVILRIALFFVGSAF
MAV_4626|M.avium_104 AAIVVVIFSMVGAEIVTVAAAESRDPRRAVQKATRSVVTRIGIFFVGSVF
Rv0346c|M.tuberculosis_H37Rv LVTSGVVFAYAAIELVGIAAGETAEPAKIMPRAINSVVLRIACFYVGSTV
Mb0354c|M.bovis_AF2122/97 LVTSGVVFAYAAIELVGIAAGETAEPAKIMPRAINSVVLRIACFYVGSTV
MMAR_0627|M.marinum_M LVTSGVVFAYAAIELVGTAAGEAAEPEKIMPRAINSVVFRIGLFYIGSTV
MLBr_01305|M.leprae_Br4923 VVSSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGSVI
MUL_0383|M.ulcerans_Agy99 VVTSGVVFAYAAVELVGTAAGETAEPEKVMPRAINSVIARIALFYVGSLF
MAB_2914|M.abscessus_ATCC_1997 IVTSGVVFAYSAVELVGIAAGETENPDKVMPRAINSVVFRVAVFYVGSLI
TH_0471|M.thermoresistible__bu AGASIVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSL
Mvan_1740|M.vanbaalenii_PYR-1 AGASIVFFAFIGFDIVATTAEETRNPQRDVPRGILASLAIVTVLYVAVAV
*.*: . :: :* *: :* : : :. : : : :::. .
Mflv_1998|M.gilvum_PYR-GCK LLVLIVPWNDESVS--ASPFVAAFNAMGIPYADHVMNAVVLTAVLSCLNS
MSMEG_5473|M.smegmatis_MC2_155 LLVTILPWNSDQTV--ASPFVAAFTEMGIPYADHIMNAVVLTAVLSCLNS
MAV_4626|M.avium_104 LLVAILPWDSVELG--ASPYVAALKHLGLAGADQVMNAVVLTAVLSCLNS
Rv0346c|M.tuberculosis_H37Rv LLALLLPYTAYKEH--VSPFVTFFSKIGIDAAGSVMNLVVLTAALSSLNA
Mb0354c|M.bovis_AF2122/97 LLALLLPYTAYKEH--VSPFVTFFSKIGIDAAGSVMNLVVLTAALSSLNA
MMAR_0627|M.marinum_M LLALLLPYTAYREH--VSPFVTFFSKVGIEGAGSVMNLVVLTAALSSLNA
MLBr_01305|M.leprae_Br4923 LLALLLPYSAFKAS--ESPFVTFFSKVGFYGAGDLMNIVVLTAALSSLNA
MUL_0383|M.ulcerans_Agy99 LLGLLLPYTDYKAG--ESPFVTFFDKIGVTGAGSIMNVVVMTAAFSSLNA
MAB_2914|M.abscessus_ATCC_1997 LLALLLPYGTYKAG--ESPFVTFFSRIGVPYAGDVMNFVVLTAALSSLNA
TH_0471|M.thermoresistible__bu VLTGMVHYTELRAAGENANLATAFAANGIDWAAKVISIGALAGLTTVVIV
Mvan_1740|M.vanbaalenii_PYR-1 VLTGMVSYTELRDA-ETQNLATAFSLNGVDWAAKVISIGALAGLTTVVIV
:* :: : .: : *. * ::. .::. : :
Mflv_1998|M.gilvum_PYR-GCK GMYTASRMLFVLAARREAPAALVKVTNRGVPANAILASSVVGFLCVIAAA
MSMEG_5473|M.smegmatis_MC2_155 GMYTASRMLFVLAARREAPPQLVAVTRRGVPATAILTSSVIGLICVIAAA
MAV_4626|M.avium_104 GLYTASRMLFVLAERGEAPARLVRLSGRGVPHIAILCSSAVGFLCVVMAR
Rv0346c|M.tuberculosis_H37Rv GLYSTGRILRSMAINGSGPRFTAPMSKTGVPYGGILLTAGIGLLGIILNA
Mb0354c|M.bovis_AF2122/97 GLYSTGRILRSMAINGSGPRFTAPMSKTGVPYGGILLTAGIGLLGIILNA
MMAR_0627|M.marinum_M GLYSTGRILRSMAINGSGPKFTAPMSKNGVPYGGILLTAGIGTLGIVLNA
MLBr_01305|M.leprae_Br4923 GLYATGRVMHSIAINGSGPKFTARMSKNGVPYGGILLAAVICLCGVALNA
MUL_0383|M.ulcerans_Agy99 GLYSTGRILRSMAMNGSAPSFTARMSQRGVPYGGICLTAAIGLIGVLLNG
MAB_2914|M.abscessus_ATCC_1997 GLYSTGRILRSLAMNGSAPKVLSRMTSGGVPFMGIAVTGALTLVGIFMNL
TH_0471|M.thermoresistible__bu LVLGQTRVLFAMSRDGLLPRPLSKTGPRGTPVR---ITVLVGALVAIAAS
Mvan_1740|M.vanbaalenii_PYR-1 LVLGQTRVLFAMSRDGLLPRQLAKTGEHGTPVR---ITLIVGAVVAVTAT
: *:: :: * *.* : :
Mflv_1998|M.gilvum_PYR-GCK VSPDTIFAFLLNSSGAIILFVYLLIAISQIVLRYRT---PDSKLR---VK
MSMEG_5473|M.smegmatis_MC2_155 FFPDTIFQFLLNSSGAVILFVYLLIAISQIVLRRRT---DDSMLK---VK
MAV_4626|M.avium_104 VAPNTVFLFLLNSSGAIILFVYWLIALSQIVLRRRT---PAERLS---VK
Rv0346c|M.tuberculosis_H37Rv IKPSQAFEIVLHIAATGVIAAWATIVACQLRLHRMA---NAGQLQRPKFR
Mb0354c|M.bovis_AF2122/97 IKPSQAFEIVLHIAATGVIAAWATIVACQLRLHRMA---NAGQLQRPKFR
MMAR_0627|M.marinum_M VKPSQAFEIVLHIAATGVIAAWATIVACQLRFHRLT---ISGVLQRPSFR
MLBr_01305|M.leprae_Br4923 FNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKMA---KAGIMRRPRFR
MUL_0383|M.ulcerans_Agy99 VVPEMAFEIVLNIAALGILASWATIVCCQIQLFRWS---KQGIVYRPSFR
MAB_2914|M.abscessus_ATCC_1997 VIPAEAFETALDLAALGIISSWATIVICQIQLYRWS---QRGILQRGSFR
TH_0471|M.thermoresistible__bu VFPVGNLEEMVN---IGTLFAFALVSAGVIVLRRTRPDLKRG--------
Mvan_1740|M.vanbaalenii_PYR-1 VFPIGKLEEMVN---IGTLFAFVLVSAGVLVLRRTRPDLKRG--------
. * : : : : : : :
Mflv_1998|M.gilvum_PYR-GCK MWLFPVLSGLTALGILAILVQMFIDT-ALRSQLVLSLLSWAVVIALFAAN
MSMEG_5473|M.smegmatis_MC2_155 MWLFPVLSILTAAAIFAILVQMFVQGGDNRSALVLSLASWAVVIVLFWLN
MAV_4626|M.avium_104 MWFFPVLSVLTLAGITAVLVQMAFDA-TARSQLWLSLLSWAVVVALY---
Rv0346c|M.tuberculosis_H37Rv MPLSPFSGYLTLAFLAGVLILMYFDEQHGPWMIAATVIGVPALIGGWYLV
Mb0354c|M.bovis_AF2122/97 MPLSPFSGYLTLAFLAGVLILMYFDEQHGPWMIAATVIGVPALIGGWYLV
MMAR_0627|M.marinum_M MPWSPYSGWITLAFLASVLILMLFDEVYGRWMLAAILVGVPALIGGWYLV
MLBr_01305|M.leprae_Br4923 MPLAPYSGYLTLAFLFAVLVVMAFDKPIGTWTVASLIVIVPALIAGWYSI
MUL_0383|M.ulcerans_Agy99 MWGAPYTGYLTLVFLFAVLVLMGFSYPIDTWTVATLGLIIPMLIGGWFLV
MAB_2914|M.abscessus_ATCC_1997 MPGAPYTGYLTLAFLAAVVVLMCYEN---AWNLIAIAVLVPVLTAGWYLC
TH_0471|M.thermoresistible__bu -FRVPFVPWLPLAAIAACLWLMLNLT---GLTWVRFLGWMALGLVLYFFY
Mvan_1740|M.vanbaalenii_PYR-1 -FRTPWVPLLPILAIAACVWLMLNLT---GLTWIRFLIWMAIGVVVYFLY
* :. : . : * . :
Mflv_1998|M.gilvum_PYR-GCK KWFVKKRPAEGDSGAPTGPPHRVLVLANRTVDSDELLAELSRIGADQTAE
MSMEG_5473|M.smegmatis_MC2_155 RMFVSRRPQTPDVAPTSEHPHRVLVLANQTVESDELLDELRRIGADRDTT
MAV_4626|M.avium_104 --FVARSRG--------------------------------RVAAR----
Rv0346c|M.tuberculosis_H37Rv RNRVTAVAH-------------------------------HAIDHTKSVA
Mb0354c|M.bovis_AF2122/97 RNRVTAVAH-------------------------------HAIDHTKSVA
MMAR_0627|M.marinum_M RNRVQVAAR-------------------------------DTVASE----
MLBr_01305|M.leprae_Br4923 RKRVMTIAR-------------------------------ERMGYTGPFP
MUL_0383|M.ulcerans_Agy99 RDRVLEIAR-------------------------------RRIGYIGEFP
MAB_2914|M.abscessus_ATCC_1997 RGRVLQLAR-------------------------------ERIGYTGAYP
TH_0471|M.thermoresistible__bu GRHHSMLGR--------------------------------GMTDISARR
Mvan_1740|M.vanbaalenii_PYR-1 GRRHSLVGR--------------------------------RMG------
:
Mflv_1998|M.gilvum_PYR-GCK YLVVVPASAVDTGAAATHGPRDVTEATQEAATARLESTLAALRGKNLTVQ
MSMEG_5473|M.smegmatis_MC2_155 YYVVVPASPIDTGVANTHGPLDISEATQQAAQERLDSTLATLRSGDFEAT
MAV_4626|M.avium_104 --------------------------------------------------
Rv0346c|M.tuberculosis_H37Rv VVHSADPI------------------------------------------
Mb0354c|M.bovis_AF2122/97 VVHSADPI------------------------------------------
MMAR_0627|M.marinum_M --------------------------------------------------
MLBr_01305|M.leprae_Br4923 AIANPPVQPSERSHSQNP--------------------------------
MUL_0383|M.ulcerans_Agy99 VVANPGPMDKK---------------------------------------
MAB_2914|M.abscessus_ATCC_1997 VIAHTPMLDDPPGESHR---------------------------------
TH_0471|M.thermoresistible__bu TG------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK GELGDYRPLRALDHAVAEFGPDQIVIATLPPEFSMWHRFDVVDRARAQFT
MSMEG_5473|M.smegmatis_MC2_155 GTLGEYRPLRALAKAVETFRPDQIVISTLPPEESVWQRFDVVDRARAEHH
MAV_4626|M.avium_104 --------------------------------------------------
Rv0346c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0354c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0627|M.marinum_M --------------------------------------------------
MLBr_01305|M.leprae_Br4923 --------------------------------------------------
MUL_0383|M.ulcerans_Agy99 --------------------------------------------------
MAB_2914|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1998|M.gilvum_PYR-GCK VPVTHVVATSAVGNQVPR
MSMEG_5473|M.smegmatis_MC2_155 LPVTHVVSRVRATEPAP-
MAV_4626|M.avium_104 ------------------
Rv0346c|M.tuberculosis_H37Rv ------------------
Mb0354c|M.bovis_AF2122/97 ------------------
MMAR_0627|M.marinum_M ------------------
MLBr_01305|M.leprae_Br4923 ------------------
MUL_0383|M.ulcerans_Agy99 ------------------
MAB_2914|M.abscessus_ATCC_1997 ------------------
TH_0471|M.thermoresistible__bu ------------------
Mvan_1740|M.vanbaalenii_PYR-1 ------------------