For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKITGTVVKLGIVSVVLLFFTVMIIVIFGQMRFDRTNGYTAEFSNVSGLRQGQFVRASGVEIGKVKALHL VDGGRRVRVEFNIDRSVPLYQSTTAQIRYSDLIGNRYVELKRGEGKGANDLLPPGGLIPLSRTSPALDLD ALIGGFKPVFRALDPAKVNNIANALITVFQGQGGTINDILDQTAQLTSQIAERDQAIGEVVKNLNIVLDT TVKHRKEFDETVNNLENLITGLRNHSDQLAGGLAHISNGAGTVADLLAENRTLVRKAVSYLDAIQQPVID QRVELDDLLHKTPTALTALGRANGTYGDFQNFYLCDLQIKWNGFQAGGPVRTVKLFSQPTGRCTPQ
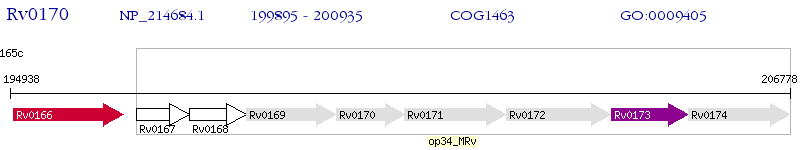
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0170 | mce1B | - | 100% (346) | MCE-FAMILY PROTEIN MCE1B |
| M. tuberculosis H37Rv | Rv0590 | mce2B | 1e-89 | 64.59% (257) | MCE-family protein MCE2B |
| M. tuberculosis H37Rv | Rv1967 | mce3B | 3e-72 | 44.15% (342) | MCE-family protein MCE3B |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0176 | mce1B | 0.0 | 99.71% (346) | MCE-family protein MCE1B |
| M. gilvum PYR-GCK | Mflv_0710 | - | 1e-129 | 64.45% (346) | virulence factor Mce family protein |
| M. leprae Br4923 | MLBr_02590 | mce1B | 1e-155 | 75.14% (346) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_4152c | - | 5e-59 | 38.36% (318) | MCE-family protein |
| M. marinum M | MMAR_0413 | mce1B | 1e-156 | 76.30% (346) | MCE-family protein Mce1B |
| M. avium 104 | MAV_5014 | - | 1e-159 | 78.03% (346) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_0135 | - | 1e-128 | 62.43% (346) | virulence factor Mce family protein |
| M. thermoresistible (build 8) | TH_3551 | - | 1e-135 | 65.90% (346) | PUTATIVE virulence factor Mce family protein |
| M. ulcerans Agy99 | MUL_1063 | mce1B | 1e-155 | 76.01% (346) | MCE-family protein Mce1B |
| M. vanbaalenii PYR-1 | Mvan_0135 | - | 1e-130 | 64.74% (346) | virulence factor Mce family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0710|M.gilvum_PYR-GCK MKITGTAIKLGAFALVLLMFTAIIIVVFGQMRFDRTTGYSAIFSNASGLR
Mvan_0135|M.vanbaalenii_PYR-1 MKITGTAIKLGAFSLVLLMFTAIIIVVFGQMRFDRTTGYSAIFSSASGLR
TH_3551|M.thermoresistible__bu MKITGTAIKLGAFSVVLLVFTALIVVVFGQVRFDRTTGYTAVFSSASGLR
MSMEG_0135|M.smegmatis_MC2_155 MSIKGTLFKLGIFSLVLLTFTALIFVVFGQIRFNRTTEYSAIFKNVSGLR
Rv0170|M.tuberculosis_H37Rv MKITGTVVKLGIVSVVLLFFTVMIIVIFGQMRFDRTNGYTAEFSNVSGLR
Mb0176|M.bovis_AF2122/97 MKITGTVVKLGIVSVVLLFFTVMIIVIFGQMRFDRTNGYTAEFSNVSGLR
MLBr_02590|M.leprae_Br4923 MKLTGTVVRLSIFSLVLLLFTVMIIVVFGQMRFGRTNGYTAEFTNISGLR
MAV_5014|M.avium_104 MKITGTLVRLSIFSVVLLIFTVMIIVVFGQMRFDRTNGYSAEFSNISGLR
MMAR_0413|M.marinum_M MKITGTAIRLGIFSLVLMLFSVMIIIVFGQIRFDRTYAYSAEFSNASGLR
MUL_1063|M.ulcerans_Agy99 MKITGTAIRLGIFSLVLMLFSVMIIIVFGQIRFDRTYAYSAEFSNASGLR
MAB_4152c|M.abscessus_ATCC_199 MKR-ATQIKVIVFTVVMLLVAAGLIITFGEFRFGSGKRYHAMFSTASDLR
*. .* .:: .::*:: .:. :.: **:.**. * * *.. *.**
Mflv_0710|M.gilvum_PYR-GCK SGQFVRASGVEVGKVKGVELIDNGSKVRVDFDVDRSLELFDETTASVRYL
Mvan_0135|M.vanbaalenii_PYR-1 PGQFVRASGVEVGKVKGVELIEGGSKVRVDFNVDRSLELFDESTASIRYL
TH_3551|M.thermoresistible__bu AGQFVRAAGVEVGKVSKVELIDGGQRVRVDFNVDRSLPLFQDTTATIRYL
MSMEG_0135|M.smegmatis_MC2_155 DGQFVRAAGVEVGKVKSVDLINGGEQAEVKFTVERSLPLFQETTAAIRYQ
Rv0170|M.tuberculosis_H37Rv QGQFVRASGVEIGKVKALHLVDGGRRVRVEFNIDRSVPLYQSTTAQIRYS
Mb0176|M.bovis_AF2122/97 QGQFVRASGVEIGKVKALHLVDGGRRVRVEFNIDRSVPLYQSTTAQIRYS
MLBr_02590|M.leprae_Br4923 TGQFVRASGVEVGKVNSVALINGGTRVRVKFNVDRSVPLYQSTTAQIRYL
MAV_5014|M.avium_104 AGQFVRASGVEVGKVSSVQLVDGGKRARVEFNVDRSVPLYQSTTAQIRYL
MMAR_0413|M.marinum_M PGQFVRASGVEIGKVKSVSLIDGGKRVRVNFDVDRSVPLYQSTSAQIRYL
MUL_1063|M.ulcerans_Agy99 PGQFVRASGVEIGKVKSVSLIDGGKRVRVNFDVDRSVPLYQSTSAQIRYL
MAB_4152c|M.abscessus_ATCC_199 SGQKVRIAGVPVGRVKNVSLNRDNS-VEVTFDVDSKYQLYSSSRAIIRYE
** ** :** :*:*. : * .. ..* * :: . *:..: * :**
Mflv_0710|M.gilvum_PYR-GCK NLIGDRYLELARGD---SNTRLAAGGTIPVERTQPALDLDALIGGFRPVF
Mvan_0135|M.vanbaalenii_PYR-1 NLIGDRYLELARGE---SNTRMAAGGTIPIERTQPALDLDALIGGFRPVF
TH_3551|M.thermoresistible__bu NLIGDRYLELKRGN---SNERLPAGGEIPLERTQPALDLDALVGGFRPLF
MSMEG_0135|M.smegmatis_MC2_155 DLIGNRYLELKRGD---SDQILPPGSTIPVERTEPALDLDALVGGFRPLF
Rv0170|M.tuberculosis_H37Rv DLIGNRYVELKRGEGKGANDLLPPGGLIPLSRTSPALDLDALIGGFKPVF
Mb0176|M.bovis_AF2122/97 DLIGNRYVELKRGEGKGANDLLPPGGLIPLSRTSPALDLDALIGGFKPVF
MLBr_02590|M.leprae_Br4923 DLIGNRYLELKHGEGQGSEKIMPAGGLIPLSRTSPALDLDALIGGFKPLF
MAV_5014|M.avium_104 DLIGNRYLELKRGQGEGADKVLPPGGFIPLSRTQPALDLDALIGGFKPLF
MMAR_0413|M.marinum_M DLIGNRYLELKRGEGDGSDRIMPPGGFIPLSRTSPALDLDALIGGFKPVF
MUL_1063|M.ulcerans_Agy99 DLIGNRYLELKRGEGDGSDRIMPPGGFIPLSRTSPALDLDALIGGFKPVF
MAB_4152c|M.abscessus_ATCC_199 NLVGDRYLEITAGPG--ELHKLPDGGTLDKEHTQPALDLDALLGGLRPVL
:*:*:**:*: * :. *. : .:*.********:**::*::
Mflv_0710|M.gilvum_PYR-GCK QSLDPEKVNTIAQSLITVFQGQGGTINDILDQTASLTSALADRDQAIGEV
Mvan_0135|M.vanbaalenii_PYR-1 QSLEPEKVNTIAQSIITVFQGQGGTINDILDQTASLTSALADRDQAIGEV
TH_3551|M.thermoresistible__bu RALDAEKVNTIAQSIITVFQGQGGTINDILDQTAQLTATLADRDQAIGEV
MSMEG_0135|M.smegmatis_MC2_155 RSLEPEKVNTIATSLITIFQGQGGTINDILDQTAQLTASLADRDQAIGEV
Rv0170|M.tuberculosis_H37Rv RALDPAKVNNIANALITVFQGQGGTINDILDQTAQLTSQIAERDQAIGEV
Mb0176|M.bovis_AF2122/97 RALDPAKVNNIANALITVFQGQGGTINDTLDQTAQLTSQIAERDQAIGEV
MLBr_02590|M.leprae_Br4923 RALDPDKINTIASAIVAAFQGQGGTINDILDQTAQLTTAIGERDQAIGEV
MAV_5014|M.avium_104 RALDPQKVNTIATALVTVFQGQGGTINDILDQTAQLTSQLGERDQAIGEV
MMAR_0413|M.marinum_M RALDPDKVNTIASALVTVFQGQGGTINDILDQTAQLTSQLAERDQAIGEV
MUL_1063|M.ulcerans_Agy99 RALDPDKVNTIASALVTVFQGQGGTINDILDQTAWLTSQLAERDQAIGEV
MAB_4152c|M.abscessus_ATCC_199 KGFDAAKVNEISNAFIQILQGQGGTLSQLLGDTSSFTSGLAERDQLIGDV
:.::. *:* *: ::: :******:.: *.:*: :*: :.:*** **:*
Mflv_0710|M.gilvum_PYR-GCK VTNLNTVLDTTVRHQQEFDDTVKNFEVLITGLKNRADPLAESVADISDAA
Mvan_0135|M.vanbaalenii_PYR-1 ITNLNTVLDTTVRHQQEFDDTVKNFEVLITGLKNRADPIATSVADISDAA
TH_3551|M.thermoresistible__bu IRNLNTVLETTVRHQKEFDETIHNFEVLITGLRNRADPIAESTANISNAA
MSMEG_0135|M.smegmatis_MC2_155 IKNLNTVLDTTVRHQKQFDETLVNFETLITGLKNRADPIATSVADISDAA
Rv0170|M.tuberculosis_H37Rv VKNLNIVLDTTVKHRKEFDETVNNLENLITGLRNHSDQLAGGLAHISNGA
Mb0176|M.bovis_AF2122/97 VKNLNIVLDTTVKHRKEFDETVNNLENLITGLRNHSDQLAGGLAHISNGA
MLBr_02590|M.leprae_Br4923 VKNLNVVLDTTVRHRKEFDQTVNNLEILITGLKDHGDQLAGSFAHISNAA
MAV_5014|M.avium_104 IKNLNTVLDTTVRHRQQFDQTINNLEVLITGLKDHGDQLAGGTAHISNAA
MMAR_0413|M.marinum_M IKNLNTVLDATVRHRKDFDQTVNNLEVLITGLNEHRDDLSGGIAHVSNAA
MUL_1063|M.ulcerans_Agy99 IKNLNTVLDATVRHRKDFDQTVNNLEVLITGLNEHRDDLSGGIAHVSNAA
MAB_4152c|M.abscessus_ATCC_199 INHLNEVLATVDSKSTQFSSSVDQLQQLISGLAQQRDVVAGALPPLASTA
: :** ** :. : :*..:: ::: **:** :: * :: . . ::. *
Mflv_0710|M.gilvum_PYR-GCK GTIGDLLADNRPLLQSTISKLEILNQPLVDQKDQLNQLLEQIPDALAIIG
Mvan_0135|M.vanbaalenii_PYR-1 GTIGDLLADNRPLLKDTVSKLEILNQPLVDQRDQLNDLLVKLPDALAIIG
TH_3551|M.thermoresistible__bu GTLADVLAQNRPLLDNSIDHLEAVQGPLVEQSAELNNILVQLPTALKTIG
MSMEG_0135|M.smegmatis_MC2_155 GSLADLLSDNRPLLKDTIGYLDVIQAPLVEQKQEVSDILVQMPQALKIIG
Rv0170|M.tuberculosis_H37Rv GTVADLLAENRTLVRKAVSYLDAIQQPVIDQRVELDDLLHKTPTALTALG
Mb0176|M.bovis_AF2122/97 GTVADLLAENRTLVRKAVSYLDAIQQPVIDQRVELDDLLHKTPTALTALG
MLBr_02590|M.leprae_Br4923 GTVANLLAEDHALLHKSINYLDGIQQPLIDQRLQLEDYLHKLPTVLNQIG
MAV_5014|M.avium_104 GTVADLLAEDRSLLHKTLNYLDAVQQPLIDQQDQLQDYLKKVPTALNMIG
MMAR_0413|M.marinum_M GTVADLLGEDRALLHKTVNYLDAVQQPLIDQRQELADFLHKVPTALNMIG
MUL_1063|M.ulcerans_Agy99 GTVADLLGEDRALLHKTVNYLDAVQQPLIDQRQELADFLHKVPTALNMIG
MAB_4152c|M.abscessus_ATCC_199 SSLESVLKDTRPYIKGTIEQARQLATRVDERKADVNVVVENLAENYLRLN
.:: .:* : :. : :: : : :: :: : : . :.
Mflv_0710|M.gilvum_PYR-GCK RSGGIYGDFFNFYACDVTLKLNGLQPGGPVRTVRVWSQPT-GRCTPQ--
Mvan_0135|M.vanbaalenii_PYR-1 RSGGVYGDFFNFYACDVTLKLNGLQPGGPVRTVKVWSQPS-GRCTPQ--
TH_3551|M.thermoresistible__bu RAGGVYGDFFNFYACDISLKLNGLQPGGPVRTVKITTQPS-GRCTPQ--
MSMEG_0135|M.smegmatis_MC2_155 RAGGIYGDFFNFYACDLTLKLNGLQPGGPVRTVRITTQPS-GRCTPK--
Rv0170|M.tuberculosis_H37Rv RANGTYGDFQNFYLCDLQIKWNGFQAGGPVRTVKLFSQPT-GRCTPQ--
Mb0176|M.bovis_AF2122/97 RANGTYGDFQNFYLCDLQIKWNGFQAGGPVRTVKLFSQPT-GRCTPQ--
MLBr_02590|M.leprae_Br4923 RTIGSYGDFVNFYSCDITLKINGLQPGGPVRTVRLFQQPT-GRCTPQ--
MAV_5014|M.avium_104 RAIGSYGDFVNFYACDITLKINGLQAGGPVRTVRLFQQPT-GRCTPQ--
MMAR_0413|M.marinum_M RAIGSYGDFVNFYSCDISLKINGLQPGGPVRTVRLFSQPT-GRCAPQ--
MUL_1063|M.ulcerans_Agy99 RAIGSYGDFVNFYSCDISLKINGLQPGGPVRTVRLFSQPT-GRCAPQ--
MAB_4152c|M.abscessus_ATCC_199 -ALGAYGSFFNIYYCSIRIKVNGPVGTDWLIPFGGPPDPSKGRCAFKNE
: * **.* *:* *.: :* ** . : .. :*: ***: :