For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AAPVVGDADLQSVRRIRLDVLGMSCAACASRVETKLNKIPGVRASVNFATRVATIDAVGMAADELCGVVE KAGYHAAPHTETTVLDKRTKDPDGAHARRLLRRLLVAAVLFVPLADLSTLFAIVPSARVPGWGYILTALA APVVTWAAWPFHSVALRNARHRTTSMETLISVGIVAATAWSLSSVFGDQPPREGSGIWRAILNSDSIYLE VAAGVTVFVLAGRYFEARAKSKAGSALRALAELGAKNVAVLLPDGAELVIPASELKKRQRFVTRPGETIA ADGVVVDGSAAIDMSAMTGEAKPVRAYPAASVVGGTVVMDGRLVIEATAVGADTQFAAMVRLVEQAQTQK ARAQRLADHIAGVFVPVVFVIAGLAGAAWLVSGAGADRAFSVTLGVLVIACPCALGLATPTAMMVASGRG AQLGIFIKGYRALETIRSIDTVVFDKTGTLTVGQLAVSTVTMAGSGTSERDREEVLGLAAAVESASEHAM AAAIVAASPDPGPVNGFVAVAGCGVSGEVGGHHVEVGKPSWITRTTPCHDAALVSARLDGESRGETVVFV SVDGVVRAALTIADTLKDSAAAAVAALRSRGLRTILLTGDNRAAADAVAAQVGIDSAVADMLPEGKVDVI QRLREEGHTVAMVGDGINDGPALVGADLGLAIGRGTDVALGAADIILVRDDLNTVPQALDLARATMRTIR MNMIWAFGYNVAAIPIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRNFGAQ*
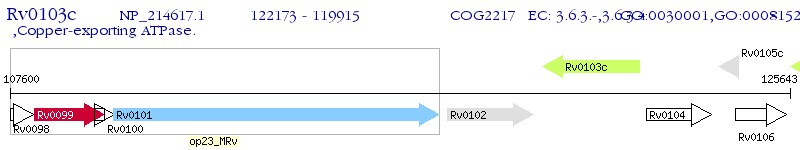
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0103c | ctpB | - | 100% (752) | PROBABLE CATION-TRANSPORTER P-TYPE ATPASE B CTPB |
| M. tuberculosis H37Rv | Rv0092 | ctpA | 0.0 | 68.15% (741) | cation transporter P-type ATPase A |
| M. tuberculosis H37Rv | Rv0969 | ctpV | e-151 | 47.12% (660) | metal cation transporter P-type ATPase CtpV |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0106c | ctpB | 0.0 | 99.87% (752) | cation-transporter P-type ATPase B |
| M. gilvum PYR-GCK | Mflv_0912 | - | 0.0 | 50.73% (751) | heavy metal translocating P-type ATPase |
| M. leprae Br4923 | MLBr_02000 | ctpB | 0.0 | 77.13% (752) | cation-transporting ATPase |
| M. abscessus ATCC 19977 | MAB_3984c | - | 1e-169 | 46.52% (748) | putative metal transporter ATPase |
| M. marinum M | MMAR_0269 | ctpB | 0.0 | 82.13% (750) | cation-transporter p-type ATPase B CtpB |
| M. avium 104 | MAV_5246 | - | 0.0 | 49.27% (755) | copper-translocating P-type ATPase |
| M. smegmatis MC2 155 | MSMEG_5014 | - | 0.0 | 50.74% (747) | copper-translocating P-type ATPase |
| M. thermoresistible (build 8) | TH_4361 | - | 0.0 | 50.86% (753) | PUTATIVE heavy metal translocating P-type ATPase |
| M. ulcerans Agy99 | MUL_4845 | ctpB | 0.0 | 81.47% (750) | cation-transporter p-type ATPase B CtpB |
| M. vanbaalenii PYR-1 | Mvan_3670 | - | 0.0 | 51.20% (748) | heavy metal translocating P-type ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0912|M.gilvum_PYR-GCK MALMT------TSTSTPVSGANVELRIGGMTCASCANRIERKLNKLDGVA
MAV_5246|M.avium_104 MSTPPRHIDEGTFPDHTASTARIELEITGMTCASCAARIEKKLNKLDGVT
MSMEG_5014|M.smegmatis_MC2_155 ---------------MTTATGHVELQIGGMTCASCAARIERKLNKLDGVT
TH_4361|M.thermoresistible__bu --------------MNTGGMNTVTLTVGGMTCASCAARVEKRLNRIDGVV
Mvan_3670|M.vanbaalenii_PYR-1 -------MSLPNHIVGVDQPHSIELSIGGMTCASCAARIEKKLNKIDGVE
MAB_3984c|M.abscessus_ATCC_199 ------------METTNTGMETTDLRLHGMTCASCASRVEKGLNKITGVN
Rv0103c|M.tuberculosis_H37Rv ----MAAPVVGDAD--LQSVRRIRLDVLGMSCAACASRVETKLNKIPGVR
Mb0106c|M.bovis_AF2122/97 ----MAAPVVGDAD--LQSVRRIRLDVSGMSCAACASRVETKLNKIPGVR
MMAR_0269|M.marinum_M ----MAAPITGDLD--LGSARRIQLDVSGMSCAACASRVETKLNKVPGVR
MUL_4845|M.ulcerans_Agy99 ----MAAPITGDLD--LGSARRIQLDVSGMSCAACASRVETKLNKVPGVR
MLBr_02000|M.leprae_Br4923 ----MTASLVEDTNNNHESVRRIQLDVAGMLCAACASRVETKLNKIPGVR
* : ** **:** *:* **:: **
Mflv_0912|M.gilvum_PYR-GCK ATVNYATEKATVTVPDGYDPALLIAEVEKTGYTATLPTPRAPAPSDASGD
MAV_5246|M.avium_104 ATVNYATEKAAVSAPASYDPQTLITEIENAGYAAAVATPSPPR-------
MSMEG_5014|M.smegmatis_MC2_155 ASVNYATEKASVEVAGEVSPETLIEVVETAGYTAHLPAPPQPETGGTEQD
TH_4361|M.thermoresistible__bu ATVNFATEQATVRYPDHVDPADLIAAIEATGYTASLPAPQRDTGGAPAGA
Mvan_3670|M.vanbaalenii_PYR-1 ASVNYATEKAKVDYPNSVDPAVLIAAVEATGYTATAPAAPNDAAPTDSGP
MAB_3984c|M.abscessus_ATCC_199 ARVNLALESARVEHDGSVTGEDLVKAVESVGYTAQVVGGSHQDAHAGDHD
Rv0103c|M.tuberculosis_H37Rv ASVNFATRVATIDAVG-MAADELCGVVEKAGYHAAPHTETTVLDKRTKDP
Mb0106c|M.bovis_AF2122/97 ASVNFATRVATIDAVG-MAADELCGVVEKAGYHAAPHTETTVLDKRTKDP
MMAR_0269|M.marinum_M ASVNFATRVATIDAVD-VAADQLCELVEKAGYQAVPHAESAADDR---DP
MUL_4845|M.ulcerans_Agy99 ASVNFATRVATIDAVD-VAADELCELVEKAGYQAVPHAESAADDR---DP
MLBr_02000|M.leprae_Br4923 ASVNFATRVATIDAVD-VAVDELRQVIEQAGYRATAHAESAVEEI---DP
* ** * . * : * :* .** *
Mflv_0912|M.gilvum_PYR-GCK TPNADGI--DDPELASLRHRLRASTVLTVPVIAMA----MIPALQFTYWQ
MAV_5246|M.avium_104 ---------DDPELASLRRRLVTAIALAGPVIAVA----MIPALQFQHWH
MSMEG_5014|M.smegmatis_MC2_155 P--------EDRATDALRTRLLVSAVLTVPVIAMA----MVPALQFTNWQ
TH_4361|M.thermoresistible__bu ATGAAGDD-HDAEAERWRRRLTISAALSIPVVVLS----MIPALQFTHWQ
Mvan_3670|M.vanbaalenii_PYR-1 DAG---------EADSLRSRLLISLVLTVPVIAMS----MIPALQFTNWQ
MAB_3984c|M.abscessus_ATCC_199 GHDPHDHMNHDVPDDQLKSRLIGSLILAIPVVVIS----MAMPLHFSGWE
Rv0103c|M.tuberculosis_H37Rv DGA---------HARRLLRRLLVAAVLFVPLADLSTLFAIVPSARVPGWG
Mb0106c|M.bovis_AF2122/97 DGA---------HARRLLRRLLVAAVLFVPLADLSTLFAIVPSARVPGWG
MMAR_0269|M.marinum_M DAE---------HARTLLRRLLVAAVLFVPLADLSTMFALVPSTRFPGWG
MUL_4845|M.ulcerans_Agy99 DAE---------HARTLLRRLLVAAVLFVPLADLSTMFALVPSTRFPGWG
MLBr_02000|M.leprae_Br4923 DAD---------YARNLLRRLIVAALLFVPLADLSTMFAIVPTNRFPGWG
** : * *: :: : . :. *
Mflv_0912|M.gilvum_PYR-GCK WASLTLAAPVIVWAAWPFHRAAWANLRHGTATMDTLISLGTVSAFLWSLY
MAV_5246|M.avium_104 WAALALTAPVVGWCGRPFHAAAWANLKHGVTTMDTLISIGTLAAFLWSLY
MSMEG_5014|M.smegmatis_MC2_155 WLSLTLAAPVVVWGALPFHRAAWLNLRHGTATMDTLISMGTLAAFGWSLY
TH_4361|M.thermoresistible__bu WLALTLTSPVVVWGAWPFHRAAVANLRHGTATMDTLISLGVTAAYLWSLW
Mvan_3670|M.vanbaalenii_PYR-1 WLALTLASPVVVWGALPFHRAAWANARHRAATMDTLISLGVSAAYLWSLW
MAB_3984c|M.abscessus_ATCC_199 ILVAALTTPILLWGGYPFHRAALVNARHGASTMDTLVSLGTIAAYLWSLW
Rv0103c|M.tuberculosis_H37Rv YILTALAAPVVTWAAWPFHSVALRNARHRTTSMETLISVGIVAATAWSLS
Mb0106c|M.bovis_AF2122/97 YILTALAAPVVTWAAWPFHSVALRNARHRTTSMETLISVGIVAATAWSLS
MMAR_0269|M.marinum_M YLLTALAAPIVTWAAWPFHSVAIRNARHRTASMETLISVGITAATVWSLS
MUL_4845|M.ulcerans_Agy99 YLLTALAAPIVIWAAWPFHSVAIRNARHRTASMETLISVGITAATVWSLS
MLBr_02000|M.leprae_Br4923 YLLTALAAPIVTWAAWPFHRVALRNARYRAASMETLISAGILAATGWSLS
:*::*:: * . *** .* * :: .::*:**:* * :* ***
Mflv_0912|M.gilvum_PYR-GCK ALFFGTAGTPGMKHPFELTLAP----SHGAA--NIYLEVAAGVTTFILAG
MAV_5246|M.avium_104 ALVLGAADRPGMRHDFELTVGHGAHVSHVAAPCHVYFEVAAGVTLFVLAG
MSMEG_5014|M.smegmatis_MC2_155 ALFWGTAGVPGMTHPFSLVGSR----TDGAG--NIYLEVAAGVTTFILAG
TH_4361|M.thermoresistible__bu ALFFTAAGMPGMTMTFTLLP-AD------TGEPHIYLEVAAAVTTFIIAG
Mvan_3670|M.vanbaalenii_PYR-1 ALFIGHAGMPGMRMAFSLLPDAG------SETEHIYLEVAAAVTVFILAG
MAB_3984c|M.abscessus_ATCC_199 VLFANAGHTHGDVH--------------------VYFEVVAAVTVFLLAG
Rv0103c|M.tuberculosis_H37Rv SVFGDQPPREGSGIWRAILNSDS-----------IYLEVAAGVTVFVLAG
Mb0106c|M.bovis_AF2122/97 SVFGDQPPREGSGIWRAILNSDS-----------IYLEVAAGVTVFVLAG
MMAR_0269|M.marinum_M TVFVDRQPRHSRGIWQAILNSDS-----------IYLEVAAGVTVFVLAG
MUL_4845|M.ulcerans_Agy99 TVFVDRQPRQSRGIWQAILNSDS-----------IYLEVAAGVTVFVLAG
MLBr_02000|M.leprae_Br4923 TIFVDKEPRQTHGIWQAILHSDS-----------IYFEVAAGVTVFVLAG
:. :*:**.*.** *::**
Mflv_0912|M.gilvum_PYR-GCK RYFEKRSKRQAGAALRALLNLGAKDVSVLR-DGVETKIAIEELMVGDEFV
MAV_5246|M.avium_104 RYFERRSKRTAGAALRALLALGAKDVAVLR-AGAETRIPIERLAVGDEFV
MSMEG_5014|M.smegmatis_MC2_155 RYFEAKSKRRASAALRALMELGAKDVAILR-NGAEQRIPIDQLGVGDEFV
TH_4361|M.thermoresistible__bu RYFEARAKRQSGAALRALLDMGANDVAVLR-DGAEKRVPIGELHVGDVFV
Mvan_3670|M.vanbaalenii_PYR-1 RYFEARAKTRSGAALRALLDMGAKDVAVVR-NGTEIRIPTAELSVGDVFV
MAB_3984c|M.abscessus_ATCC_199 RFAESRAKRSAGSALRALLSLGAKDVAVLR-GGHETRIPVDQLAVGDEFV
Rv0103c|M.tuberculosis_H37Rv RYFEARAKSKAGSALRALAELGAKNVAVLLPDGAELVIPASELKKRQRFV
Mb0106c|M.bovis_AF2122/97 RYFEARAKSKAGSALRALAELGAKNVAVLLPDGAELVIPASELKKRQRFV
MMAR_0269|M.marinum_M RYFEARAKSKAGGALRALAALGAKDVTVLLPDGAELVIPAAELKKRQRFV
MUL_4845|M.ulcerans_Agy99 RYFEARAKSKAGGALRALAALGAKDVTVLLPDGAELVIPAAELKKRQRFV
MLBr_02000|M.leprae_Br4923 RFFEARAKSKAGSALRALAARGAKNVEVLLPNGAELTIPAGELKKQQHFL
*: * ::* :..***** **::* :: * * :. .* : *:
Mflv_0912|M.gilvum_PYR-GCK VRPGEKIATDGVVTSGSSAVDASMLTGESVPVEVAAGDTVVGATVNAGGR
MAV_5246|M.avium_104 VRPGERIATDGIVVAGSSAVDAAMLTGESVPVEVGVGDGVTGGTVNAGGR
MSMEG_5014|M.smegmatis_MC2_155 VRPGEKIATDGVVVDGSSAVDASMLTGESVPVEVGPGDQVVGATVNVGGR
TH_4361|M.thermoresistible__bu VRPGEKIATDGEVVAGNSAVDASMLTGESVPVEVGPGDQVVGATVNVGGH
Mvan_3670|M.vanbaalenii_PYR-1 VRPGEKIATDGVVTDGSSAVDASMLTGESVPVEVRPGDHVTGATVNAGGR
MAB_3984c|M.abscessus_ATCC_199 VRPGERVATDGEVIEGHSALDTSTMTGESVPMEVTVGGPVLGGYVNTHGR
Rv0103c|M.tuberculosis_H37Rv TRPGETIAADGVVVDGSAAIDMSAMTGEAKPVRAYPAASVVGGTVVMDGR
Mb0106c|M.bovis_AF2122/97 TRPGETIAADGVVVDGSAAIDMSAMTGEAKPVRAYPAASVVGGTVVMDGR
MMAR_0269|M.marinum_M TRPGQTIAADGVVIDGTAAIDMSAMTGESKPVRATADAPVIGGTIVLDGR
MUL_4845|M.ulcerans_Agy99 TRPGQTIAADGVVIDGTAAIDMSAMTGESKPVRATADAPVIGGTIVLDGR
MLBr_02000|M.leprae_Br4923 VRPGETITADGVVIDGTATIDMSAITGEARPVHASPASTVVGGTTVLDGR
.***: :::** * * :::* : :***: *:.. * *. *:
Mflv_0912|M.gilvum_PYR-GCK LVVRASRVGSDTQLAQMAELVERAQTGKAEVQRLADRISGVFVPIVIAIA
MAV_5246|M.avium_104 LVVRATGIGDDTQLARMAQLVERAQSGKADAQRLADRVSGVFVPVVLLLA
MSMEG_5014|M.smegmatis_MC2_155 LVVRANRIGSDTQLAQMARLVEDAQSGKAQAQRLADRVSAVFVPIVIALA
TH_4361|M.thermoresistible__bu LQVRATRVGADTQLAQMARLVTEAQSGKAEVQRLADRVSAVFVPIVIALS
Mvan_3670|M.vanbaalenii_PYR-1 LLVRATRIGADTQLAQMARLVEDAQNGKASVQRLADRVSAVFVPIVLALS
MAB_3984c|M.abscessus_ATCC_199 IVVRANKVGADTQLARMARLVSDAQTGKAQVQRLADRVSSVFVPVVLAIS
Rv0103c|M.tuberculosis_H37Rv LVIEATAVGADTQFAAMVRLVEQAQTQKARAQRLADHIAGVFVPVVFVIA
Mb0106c|M.bovis_AF2122/97 LVIEATAVGADTQFAAMVRLVEQAQTQKARAQRLADHIAGVFVPVVFVIA
MMAR_0269|M.marinum_M LVIEATAVGADTQFAAMVRLVEEAQAQKARAQRLADRISAVFVPVVFAIA
MUL_4845|M.ulcerans_Agy99 LVIEATAVGADTQFAAMVRLVEEAQAQKARAQRLADRISAVFVPVVFAIA
MLBr_02000|M.leprae_Br4923 LVIEATAVGGDTQFAAMVRLVEDAQVQKARVQHLADRIAAVFVPMVFVIA
: :.*. :* ***:* *..** ** ** .*:***:::.****:*: ::
Mflv_0912|M.gilvum_PYR-GCK VITLGAWLGAGFS-VTAAFTAAVAVLVIACPCALGLATPTALLVGTGRGA
MAV_5246|M.avium_104 VATLAGWLAAGGT-LATALTAAVAVLIIACPCALGLATPTALLVGTGRAA
MSMEG_5014|M.smegmatis_MC2_155 VATLGFWLGTGGS-VAAAFTAAVAVLIIACPCALGLATPTALLVGTGRGA
TH_4361|M.thermoresistible__bu ALTLAGWLLLTDAPVSAAFTAAVAVLIIACPCALGLATPTALLVGTGRGA
Mvan_3670|M.vanbaalenii_PYR-1 VATLAAWLLTGHS-ATVAFTAAVAVLIIACPCALGLATPTALMVGTGRGA
MAB_3984c|M.abscessus_ATCC_199 ALTFVAWAALGNS-LASAFTAAVAVLIIACPCALGLATPTAILVGTGRGA
Rv0103c|M.tuberculosis_H37Rv GLAGAAWLVSGAG-ADRAFSVTLGVLVIACPCALGLATPTAMMVASGRGA
Mb0106c|M.bovis_AF2122/97 GLAGAAWLVSGAG-ADRAFSVTLGVLVIACPCALGLATPTAMMVASGRGA
MMAR_0269|M.marinum_M GLAGAAWLLSGAG-ADRAFSVTLGVLVIACPCALGLATPTAMMVASGRGA
MUL_4845|M.ulcerans_Agy99 GVAGVAWMLSGAG-ADRAFSVTLGVLVIACPCALGLATPTAMMVASGRGA
MLBr_02000|M.leprae_Br4923 GLAGASWLLAGAS-PDRAFSVVLGVLVIACPCTLGLATPTAMMVASGRGA
: * *::..:.**:*****:********::*.:**.*
Mflv_0912|M.gilvum_PYR-GCK QMGVLIKGPEVLESTRKVDTVVLDKTGTVTTGKMALVDVIAAPGT----E
MAV_5246|M.avium_104 QLGVLIKGPEVLETTRAVDTVVLDKTGTVTTGAMTVLDVVAADGT----D
MSMEG_5014|M.smegmatis_MC2_155 QLGILIKGPEVLESTRRVDTIVLDKTGTITTGSMTLLDVVVADGE----D
TH_4361|M.thermoresistible__bu QLGILIKGPQVLESTRRIDTVVLDKTGTVTTGQMSVTSVHVADGL----S
Mvan_3670|M.vanbaalenii_PYR-1 QLGILIKGPQVLESTRRVDTIVLDKTGTVTTGRMSLVGVHPGAGQ----N
MAB_3984c|M.abscessus_ATCC_199 QLGVLIKGPEVLENVKDIDTVVLDKTGTVTTGVMSVVEVRRSIGSGV-AD
Rv0103c|M.tuberculosis_H37Rv QLGIFIKGYRALETIRSIDTVVFDKTGTLTVGQLAVSTVTMAGSGTSERD
Mb0106c|M.bovis_AF2122/97 QLGIFIKGYRALETIRSIDTVVFDKTGTLTVGQLAVSTVTMAGSGTSERD
MMAR_0269|M.marinum_M QLGIFIKGYQALETIHGIDAVVFDKTGTLTVGKLTVNTVTTTGG----RV
MUL_4845|M.ulcerans_Agy99 QLGIFIKGYQALETIHGIDAVVFEKTGTLTVGKLTVNTVTTTGG----RV
MLBr_02000|M.leprae_Br4923 QLGIFIKGYRALETINAIDTVVFDKTGTLTLGQLSVSTVTSTGG----WC
*:*::*** ..**. . :*::*::****:* * ::: * .
Mflv_0912|M.gilvum_PYR-GCK RAELLRLAGALENASEHPIAHAIATAAAEEVGTLPTPEGFANVEGKGVQG
MAV_5246|M.avium_104 RATLLRYAGALEAASEHPIAHAIARDAKAELGPLPTPTGFRAVGGGGVHG
MSMEG_5014|M.smegmatis_MC2_155 RDDVLRVAAAVEDASEHPIARAIAAGARERTGELPAVEGFANIEGLGVQG
TH_4361|M.thermoresistible__bu ETELLRLAGAVEHASEHPIARAVATHAAQTVPPLPEVSDFENHGGNGVSG
Mvan_3670|M.vanbaalenii_PYR-1 SDELLEFAGALESASQHPIAEAIAASAAAGG-TLPAVEDFSNHNGYGVTG
MAB_3984c|M.abscessus_ATCC_199 ADEILRIAASVESASEHPVARAIVAAAASARLTLSPVTNFRNEPGVGVSG
Rv0103c|M.tuberculosis_H37Rv REEVLGLAAAVESASEHAMAAAIVAASPDPG----PVNGFVAVAGCGVSG
Mb0106c|M.bovis_AF2122/97 REEVLGLAAAVESASEHAMAAAIVAASPDPG----PVNGFVAVAGCGVSG
MMAR_0269|M.marinum_M REEVLALASAVEAASEHAVATAIVAASPHPA----PVTDFVAVAGCGVSG
MUL_4845|M.ulcerans_Agy99 REEVLALASAVEAASEHAVATAIVAASPHPA----PVTDFVAVAGCGVSR
MLBr_02000|M.leprae_Br4923 SGEVLALASAVEAASEHSVATAIVAAYADPR----PVADFVAFAGCGVSG
:* *.::* **:*.:* *:. .* * **
Mflv_0912|M.gilvum_PYR-GCK VVDGHAVVVGRESLLAD-WAQHLSPDLSRAKAGAEAQGKTVVAVGWDGQA
MAV_5246|M.avium_104 RVDGHAVAIGRPSWLAE-RGLRPDAALAAAAARAEHDGKTVVAVGWDGRA
MSMEG_5014|M.smegmatis_MC2_155 TVEGRAVVVGRPRLFGD-PEKSLPRKLADAMEAAQAQGRTAVVVGWDGTA
TH_4361|M.thermoresistible__bu RVDGHDVRVGRLAWLG----LSAGDRLRNAAAQAEAAGHTPVWVAVDGRI
Mvan_3670|M.vanbaalenii_PYR-1 VVAGHAVLVGRLSWLTDDWSLTAPDDLTSAAEHAQSQGHTPVWVAWDGAV
MAB_3984c|M.abscessus_ATCC_199 VVDGQEVSI-----------------------EAIEGQNTTAAVSWGGLR
Rv0103c|M.tuberculosis_H37Rv EVGGHHVEVGKPSWITR-TTPCHDAALVSARLDGESRGETVVFVSVDGVV
Mb0106c|M.bovis_AF2122/97 EVGGHHVEVGKPSWITR-TTPCHDAALVSARLDGESRGETVVFVSVDGVV
MMAR_0269|M.marinum_M IVDGHRIEVGKPSWITR-NT-AADEVLDTARAQGESRGETIVFVSVDGEV
MUL_4845|M.ulcerans_Agy99 IVDGHRIEVGKPSWITR-NT-AADEVLDTARAQGESRGETIVFVSVDGEV
MLBr_02000|M.leprae_Br4923 VVAEHHVKIGKPSWVTR-NA-PCDVVLESARREGESRGETVVFVSVDGVA
* : : : . .* . *. .*
Mflv_0912|M.gilvum_PYR-GCK RGVLVVADTVKPTSAQAISQMRDLGLTPVLLTGDNEAVARQIAAEVGID-
MAV_5246|M.avium_104 RGILALADTVKPSSAAAVRQFTRLGLTPILLTGDNHTVARRTAGELGIG-
MSMEG_5014|M.smegmatis_MC2_155 RGVLVVADAVKATSAEAIGQLRKLGLQPIMLTGDNGAAARAIADQVGID-
TH_4361|M.thermoresistible__bu AGVIVVSDTVKDHAAEAIAELRALGATPVLLTGDNDRAARTVAAQVGID-
Mvan_3670|M.vanbaalenii_PYR-1 RGVIVVADTVKGTSADAIGQFRKLGLRPFLLTGDNRRAALAVAAEVGIDP
MAB_3984c|M.abscessus_ATCC_199 RGEIEVADTVKPSSRRAIDELKAMGITPVLLTGDNAEVAARVASEVGIAA
Rv0103c|M.tuberculosis_H37Rv RAALTIADTLKDSAAAAVAALRSRGLRTILLTGDNRAAADAVAAQVGID-
Mb0106c|M.bovis_AF2122/97 RAALTIADTLKDSAAAAVAALRSRGLRTILLTGDNRAAADAVAAQVGID-
MMAR_0269|M.marinum_M CAAVTISDTVKDSAADAIAALRSRGLRTILLTGDNQAAADSVATQLGID-
MUL_4845|M.ulcerans_Agy99 CAAVTISDTVKDSAADAIAALRSRGLRTILLTGDNQAVADSVATQLGID-
MLBr_02000|M.leprae_Br4923 CGAVAIADTVKDSAADAISALCSRGLHTILLTGDNQAAARAVAAQVGID-
. : ::*::* : *: : * ..:***** .* * ::**
Mflv_0912|M.gilvum_PYR-GCK -TVIAEVMPKDKVDVIARLQSEGKTVAMVGDGVNDAPALAQADLGLAMGT
MAV_5246|M.avium_104 -EVISGALPADKVEAVKRLQSAGRVVAMVGDGVNDAAALAAADLGLAMGT
MSMEG_5014|M.smegmatis_MC2_155 -DVYAEVLPQDKADVIKRLQAEGRVVAMVGDGVNDAAALAQADLGLSMGT
TH_4361|M.thermoresistible__bu -DVIAGVLPAEKVDHIKQLQDRGRVVAMIGDGVNDAAALAQADLGLAIGT
Mvan_3670|M.vanbaalenii_PYR-1 DDVIAEVLPADKVDTIKCLQAQGRTVAMVGDGVNDAAALVQADLGLAMGT
MAB_3984c|M.abscessus_ATCC_199 GDVIAGVKPTEKADVVKRLQAEGKRVAMVGDGVNDSAALATADIGMAMGT
Rv0103c|M.tuberculosis_H37Rv -SAVADMLPEGKVDVIQRLREEGHTVAMVGDGINDGPALVGADLGLAIGR
Mb0106c|M.bovis_AF2122/97 -SAVADMLPEGKVDVIQRLREEGHTVAMVGDGINDGPALVGADLGLAIGR
MMAR_0269|M.marinum_M -TAIADVLPDGKVNVIRQLRDQGHTVAMVGDGINDGPALVSADLGLAIGR
MUL_4845|M.ulcerans_Agy99 -TAIADVLPDGKVNVIRQLRDQGHTVAMVGDGINDGPALVSADLGLAIGR
MLBr_02000|M.leprae_Br4923 -TVIADMLPEAKVDVIQRLRDQGHTVAMVGDGINDGPALACADLGLAMGR
. : * *.: : *: *: ***:***:**..**. **:*:::*
Mflv_0912|M.gilvum_PYR-GCK GTDVAIEASDITLVRGDLRSAVDAIRLSRRTLSTIKTNLFWAFAYNVAAI
MAV_5246|M.avium_104 GTDVAIEAADVTVVRGDLRAAVDAIRLSRRTLATIKTNLVWAFGYNLAAI
MSMEG_5014|M.smegmatis_MC2_155 GTDVAIEAGDLTLVRGDLRTAADAIRLSRRTLATIKGNLFWAFAYNVAAL
TH_4361|M.thermoresistible__bu GTDVAIEASDLTLVTGDLRAAPDAIRLGRATLRTIKGNLFWAFAYNVAAL
Mvan_3670|M.vanbaalenii_PYR-1 GSDVAIEASDLTLVRGDLRAAADAIRLARSTLRTIKGNLFWAFAYNVAAL
MAB_3984c|M.abscessus_ATCC_199 GTDAAIEASDLTLVRGDLTTVPTALRLSRKTLGTIKANLFWAFAYNTAAI
Rv0103c|M.tuberculosis_H37Rv GTDVALGAADIILVRDDLNTVPQALDLARATMRTIRMNMIWAFGYNVAAI
Mb0106c|M.bovis_AF2122/97 GTDVALGAADIILVRDDLNTVPQALDLARATMRTIRMNMIWAFGYNVAAI
MMAR_0269|M.marinum_M GTDVAIGAADIILVRDDLHIVGQALDLARATLRTIRTNMIWAFGYNVAAI
MUL_4845|M.ulcerans_Agy99 GTDVAIGAADIILVRDDLHIVGQALDLARATMRTIRTNMIWAFGYNVAAI
MLBr_02000|M.leprae_Br4923 GTDVAIGAADLILVRDSLGVVPVALDLARATMRTIRINMIWAFGYNVAAI
*:*.*: *.*: :* ..* . *: *.* *: **: *:.***.** **:
Mflv_0912|M.gilvum_PYR-GCK PVAALGMLNPMLAGAAMAFSSVFVVGNSLRLRGFTSTSSAPTH-------
MAV_5246|M.avium_104 PLAALGMLNPMLAGAAMALSSVLVVGNSLRLRSFASIIPGA---------
MSMEG_5014|M.smegmatis_MC2_155 PLAAAGLLNPMLAGAAMAFSSVFVVSNSLRLKRFR---------------
TH_4361|M.thermoresistible__bu PLAAAGLLNPLIAGAAMAFSSVFVVSNSLRLRRFTPSR------------
Mvan_3670|M.vanbaalenii_PYR-1 PLAALGLLNPLIAGAAMAFSSVFVVSNSLRLRRFTSSASPGQTEDVHVAT
MAB_3984c|M.abscessus_ATCC_199 PLAALGLLNPMIAGAAMAMSSVFVVLNSLRLRTFR---------------
Rv0103c|M.tuberculosis_H37Rv PIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRNFGAQ-------------
Mb0106c|M.bovis_AF2122/97 PIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRNFGAQ-------------
MMAR_0269|M.marinum_M PIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRNFGPPTARIVASNGEIDD
MUL_4845|M.ulcerans_Agy99 PIAAAGLLNPLIAGAAMAFSSFFVVSNSLRLRNFGPPTARIVASNGEIDD
MLBr_02000|M.leprae_Br4923 PIASSGLLNPLIAGAAMAFSSFFVVSNSLRLSNFGLSQTSD---------
*:*: *:***::******:**.:** ***** *
Mflv_0912|M.gilvum_PYR-GCK ---------------
MAV_5246|M.avium_104 ---------------
MSMEG_5014|M.smegmatis_MC2_155 ---------------
TH_4361|M.thermoresistible__bu ---------------
Mvan_3670|M.vanbaalenii_PYR-1 VPPGGDEYRRAHTAP
MAB_3984c|M.abscessus_ATCC_199 ---------------
Rv0103c|M.tuberculosis_H37Rv ---------------
Mb0106c|M.bovis_AF2122/97 ---------------
MMAR_0269|M.marinum_M ---------------
MUL_4845|M.ulcerans_Agy99 ---------------
MLBr_02000|M.leprae_Br4923 ---------------