For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSYLAGAAQIGGVMVGAPLVIGMTRQVRARWEGRAGAGLLQPWRDLLKQLGKQQITPAGTTIVFAAAPVI VAGTTLLIAAIAPLVATGSPLDPSADLFAVVGLLFLGTVALTLAGIDTGTSFGGMGASREITIAALVEPT ILLAVFALSIPAGSANLGALVASTIDHPGHVVSLAGVLAFVALVIVIVAETGRLPVDNPATHLELTMVHE AMVLEYAGPRLALVEWAAGMRLTVLLALLANLFLPWGIAGAAPTALDVLTGVVAVAAKVAILAVLLATFE VFLAKLRLFRVPELLAGSFLLALLAVTAANFFTVGA
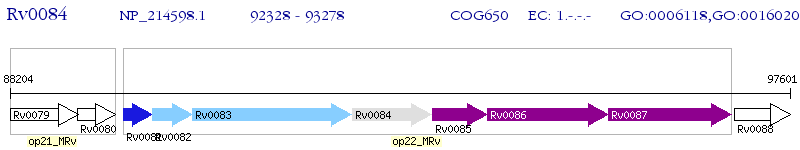
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0084 | hycD | - | 100% (316) | POSSIBLE FORMATE HYDROGENLYASE HYCD (FHL) |
| M. tuberculosis H37Rv | Rv3152 | nuoH | 7e-10 | 28.44% (225) | NADH dehydrogenase subunit H |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0087 | hycD | 1e-171 | 100.00% (316) | formate hydrogenlyase HYCD |
| M. gilvum PYR-GCK | Mflv_4488 | - | 2e-09 | 26.72% (247) | NADH dehydrogenase subunit H |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2141 | - | 1e-09 | 26.24% (263) | NADH dehydrogenase subunit H |
| M. marinum M | MMAR_1656 | hycD | 1e-141 | 81.15% (313) | formate hydrogenlyase HycD |
| M. avium 104 | MAV_5112 | - | 1e-148 | 86.39% (316) | hydrogenase-4, C subunit, putative |
| M. smegmatis MC2 155 | MSMEG_2056 | nuoH | 1e-09 | 27.85% (219) | NADH dehydrogenase subunit H |
| M. thermoresistible (build 8) | TH_1086 | nuoH | 3e-08 | 25.20% (246) | PROBABLE NADH DEHYDROGENASE I (CHAIN H) NUOH |
| M. ulcerans Agy99 | MUL_1893 | hycD | 1e-140 | 80.83% (313) | formate hydrogenlyase HycD |
| M. vanbaalenii PYR-1 | Mvan_1878 | - | 2e-10 | 27.13% (247) | NADH dehydrogenase subunit H |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4488|M.gilvum_PYR-GCK MT-YPDPTLFGHDPWWLILAKSLGVFVFLLLTVLAAILIERKILGRMQLR
Mvan_1878|M.vanbaalenii_PYR-1 MT-YPDPTLFGHDPWWLVLAKSLGIFAFLLLTVLAAILIERKVLGRMQLR
MSMEG_2056|M.smegmatis_MC2_155 MT-HPDPTLFGHDPWWLMLAKAVAIFVFLLLTVLSAILIERKLLGRMQMR
TH_1086|M.thermoresistible__bu VT-YPDPGAFGHDPWWLIVAKTVGIFGFLVLTVLSAILIERKILGRMQMR
MAB_2141|M.abscessus_ATCC_1997 MTGRTDLSQFGIDPLWLVLVKCVAVFAFLVLTVLIAILAERRVLAWMQRR
Rv0084|M.tuberculosis_H37Rv ----------------MSYLAGAAQIGGVMVGAPLVIGMTRQVRARWEGR
Mb0087|M.bovis_AF2122/97 ----------------MSYLAGAAQIGGVMVGAPLVIGMTRQVRARWEGR
MAV_5112|M.avium_104 -------------MNVMSYVAGAAQILAVMAGAPLVIGAMRQVRARLEGR
MMAR_1656|M.marinum_M -------------MNLMSAMAGAAQIGAVILGAPLVIGLMRQIRARSEGR
MUL_1893|M.ulcerans_Agy99 ----------------MSAMAGAAQIGAVILGAPLVIGLMRQIRARSEGR
: . : :: . .* *:: . : *
Mflv_4488|M.gilvum_PYR-GCK LGPNRVGPRGLLQSLADGIKLALKEGLVPAGVDKWIYLAAPVISVIPAFM
Mvan_1878|M.vanbaalenii_PYR-1 FGPNRVGPHGLLQSLADGVKLALKEGLVPAGVDKWIYLAAPIISVIPAFM
MSMEG_2056|M.smegmatis_MC2_155 FGPNRVGPAGLLQSLADGIKLALKEGLVPAGVDKPIYLLAPVISVIPAFV
TH_1086|M.thermoresistible__bu FGPNRVGPRGLLQSVADGVKLALKEGLIPRAADKPLYLLAPVIAVIPAFM
MAB_2141|M.abscessus_ATCC_1997 IGPNRVGPFGLLQSLADGVKLALKEGLTPAGVDKPIYLLAPIISVVPAIV
Rv0084|M.tuberculosis_H37Rv AGA------GLLQPWRDLLKQLGKQQITPAGT-------TIVFAAAPVIV
Mb0087|M.bovis_AF2122/97 AGA------GLLQPWRDLLKQLGKQQITPAGT-------TIVFAAAPVIV
MAV_5112|M.avium_104 GGG------GVLQPWRDLRKQLGKQQISPQGT-------TVVFAAAPAIV
MMAR_1656|M.marinum_M CGA------GILQPWRDLRKQLRKQQVTPEGT-------TAVFAVAPAVV
MUL_1893|M.ulcerans_Agy99 CGA------GILQPWRDLHKQLRKQQVTPEGT-------TAVFAVAPAVV
* *:**. * * *: : * .. : :::. *..:
Mflv_4488|M.gilvum_PYR-GCK AFAVIPLGPEVSVFGHRTALQLTDLPVAVLYILAVTSIGVYGIVLAGWAS
Mvan_1878|M.vanbaalenii_PYR-1 AFAVIPMGGEVSVFGHRTALQLTDLPVAVLYILAVTSIGVYGIVLAGWAS
MSMEG_2056|M.smegmatis_MC2_155 AFSVIPLGGAVSVFGHRTPLQLTDLPVAVLFILAATSIGVYGIVLAGWAS
TH_1086|M.thermoresistible__bu AFAVIPMGPVVSVFGHRTPLQLTDLPVAVLYILAVASVGVYGFILAGWSS
MAB_2141|M.abscessus_ATCC_1997 AYAVIPFGPVVSVFGHRTPLQLTDLPVAILFVLAVTSVGVYGIVLGGWAS
Rv0084|M.tuberculosis_H37Rv AGTTLLIAAIAPLVATGSPLDP---SADLFAVVGLLFLGTVALTLAGIDT
Mb0087|M.bovis_AF2122/97 AGTTLLIAAIAPLVATGSPLDP---SADLFAVVGLLFLGTVALTLAGIDT
MAV_5112|M.avium_104 AGTTLLIVAIAPIVATGSPLDA---AADLFAVVGLLFLGTVALTLAGIDT
MMAR_1656|M.marinum_M AATTLLIAAIAPLVATGSPLDP---VGDLFVVVGLLFLGTVALTLAGIDT
MUL_1893|M.ulcerans_Agy99 AATTLLIAAIAPLVATGSPLDP---VGDLFVVVGLLFLGTVALTLAGIDT
* :.: : ..:.. :.*: :: ::. :*. .: *.* :
Mflv_4488|M.gilvum_PYR-GCK GSVYPLLGGLRSSAQVISYEIAMALSFVAVFIYAGTMSTSGIVAAQNDVW
Mvan_1878|M.vanbaalenii_PYR-1 GSVYPLLGGLRSSAQVISYEIAMALSFAAVFIYAGTMSTSGIVAAQNDTW
MSMEG_2056|M.smegmatis_MC2_155 GSTYPLLGGLRSSAQVVSYEIAMGLSFVAVFLYAGTMSTSGIVAAQDRTW
TH_1086|M.thermoresistible__bu GSTYPLLGGLRSTAQVISYEVAMALTFVAVFLYAGTMSTSGIVAAQQGTW
MAB_2141|M.abscessus_ATCC_1997 GSTYPLLGGLRSSAQVISYEIAMGLTFAAVFLLAGTMSTSGIVAAQEGRW
Rv0084|M.tuberculosis_H37Rv GTSFGGMGASREITIAALVEPTILLAVFALSIPAGSANLGALVASTIDHP
Mb0087|M.bovis_AF2122/97 GTSFGGMGASREITIAALVEPTILLAVFALSIPAGSANLGALVASTIDHP
MAV_5112|M.avium_104 GTSFGGMGASREITIAALVEPTILLAVFALSIPAGSANLGALVTNTIDHP
MMAR_1656|M.marinum_M GTSFGGMGASREITIAALVEPTILLAVFALSIPAKSANLGAIVAFSLENP
MUL_1893|M.ulcerans_Agy99 GTSFGGMGASREITIAALVEPTILLAVFALSIPAKSANLGAIVAFSLENP
*: : :*. *. : . * :: *:. *: : * : . ..:*:
Mflv_4488|M.gilvum_PYR-GCK FIFLLLP---SFLVYLTSMVGETNRAPFDLPEAEGELVG---GFHTEYSS
Mvan_1878|M.vanbaalenii_PYR-1 YIVLLLP---SFLVYVTAMVGETNRAPFDLPEAEGELVG---GFHTEYSS
MSMEG_2056|M.smegmatis_MC2_155 FVFLLLP---SFLVYVVSMVGETNRAPFDLPEAEGELVG---GFHTEYSS
TH_1086|M.thermoresistible__bu YLFLLLP---SFLVFLVAMVGETNRAPFDLPEAEGELVG---GFHTEYSS
MAB_2141|M.abscessus_ATCC_1997 YLFLLLP---SFLVYVTAMVGETNRAPFDLPEAEGELVG---GFHTEYSS
Rv0084|M.tuberculosis_H37Rv GHVVSLAGVLAFVALVIVIVAETGRLPVDNPATHLELTMVHEAMVLEYAG
Mb0087|M.bovis_AF2122/97 GHVVSLAGVLAFVALVIVIVAETGRLPVDNPATHLELTMVHEAMVLEYAG
MAV_5112|M.avium_104 GQVVSLTGLLAFVALVIVIVAETGRLPVDNPATHLELTMVHEAMVLEYAG
MMAR_1656|M.marinum_M AEVVSLAGILAFVALVIVVIAETGRLPVDNPATHLELTMVHEAMVLEYAG
MUL_1893|M.ulcerans_Agy99 AEVVSLAGILAFVALVIVVIAETGRLSVDNPATHLELTMVHEAMVLEYAG
.: *. :*:. : ::.**.* ..* * :. **. .: **:.
Mflv_4488|M.gilvum_PYR-GCK LKFAMFMLAEYVNMTTVSALATTLFLGGWHAPWPISIADGANSDWWPLLW
Mvan_1878|M.vanbaalenii_PYR-1 LKFAMFMLAEYVNMTTVSALATTLFLGGWHAPWPISIADGANSGWWPLLW
MSMEG_2056|M.smegmatis_MC2_155 LKFAMFMLAEYVNMTTVSALATTMFLGGWHAPFPFNLIDGANSGWWPLLW
TH_1086|M.thermoresistible__bu LKFAMFMLAEYINMATVSALATTLFLGGWAAPWPINMWAGANSGWWPLLW
MAB_2141|M.abscessus_ATCC_1997 LRFAMFMLAEYINMATVSGLAATMFLGGWHAPWPLSLIDGANTSWLPALW
Rv0084|M.tuberculosis_H37Rv PRLALVEWAAGMRLTVLLALLANLFLP-WGIAGAAPTALDVLTGVVAVAA
Mb0087|M.bovis_AF2122/97 PRLALVEWAAGMRLTVLLALLANLFLP-WGIAGAAPTALDVLTGVVAVAA
MAV_5112|M.avium_104 PRLALVEWASGMRLTVLLALLANLFLP-WGIAGGHPSAVDVAIGLAAITA
MMAR_1656|M.marinum_M PRLGVIEWASGMRLTVLLALLANLFVP-WGIAGADPSLIGIAVGIAAISI
MUL_1893|M.ulcerans_Agy99 PRLGVIEWASGMRLTVLLALLANLFVP-WGIAGADPSLIGIAVGIAAISI
::.:. * :.::.: .* :.:*: * . . . .
Mflv_4488|M.gilvum_PYR-GCK FTAKVWLFLFFFMWLRATLPRMRYDQFMRLGWKLLIPVSLAWIAIVATTR
Mvan_1878|M.vanbaalenii_PYR-1 FTVKVWLFLFFFMWLRATLPRMRYDQFMALGWKILIPVSLGWIMIVAITH
MSMEG_2056|M.smegmatis_MC2_155 FTAKVWTFMFLYFWLRATLPRLRYDQFMALGWKVLIPVSLLWIMVVAITR
TH_1086|M.thermoresistible__bu FTAKVWVFLFVFFWLRATLPRLRYDQFMALGWKVLIPMSMVWIMVVATAR
MAB_2141|M.abscessus_ATCC_1997 FVAKVWGFMFLFMWLRATLPRLRYDQFMRLGWEVLIPVALVWIVVVGVVK
Rv0084|M.tuberculosis_H37Rv KVAILAVLLATFEVFLAKLRLFRVPELLAGSFLLALLAVTAANFFTVGA-
Mb0087|M.bovis_AF2122/97 KVAILAVLLATFEVFLAKLRLFRVPELLAGSFLLALLAVTAANFFTVGA-
MAV_5112|M.avium_104 KVTVLAVALAGAEVFIAKLRLFRVPELLAGSFLLALLAVTAANFFTARA-
MMAR_1656|M.marinum_M KVAIAAGVLATVEVFIAKLRLFRVPELLAGSFLLALLAVTSANFFTAQV-
MUL_1893|M.ulcerans_Agy99 KVAIAAGVLATVEVFIAKLRLFRVPELLAGSFLLALLAVTSANFFTAQV-
.. : : *.* :* ::: .: : : .. .
Mflv_4488|M.gilvum_PYR-GCK AMQNQGYQGWVTALIGVAGVAAILASLLAWRALRARRRRTS-HSPPAQSS
Mvan_1878|M.vanbaalenii_PYR-1 SLRDQGYQAPATAAIGLAVAAVILLALLGRSRLRARRIRP----EPIRPA
MSMEG_2056|M.smegmatis_MC2_155 SLRQHGEGTWAAWLLTAAVVVVVALIWGLATSLRRRTVQP----PPPQST
TH_1086|M.thermoresistible__bu GLQVFGYVGWSTGLTSVAAITALALIAVVWWVFSRRNEAE----PALPQT
MAB_2141|M.abscessus_ATCC_1997 AMGLYGYGN-PSIILGITGVAVSLGSIGLVGVLSRRTQPPKPVLPARFDP
Rv0084|M.tuberculosis_H37Rv --------------------------------------------------
Mb0087|M.bovis_AF2122/97 --------------------------------------------------
MAV_5112|M.avium_104 --------------------------------------------------
MMAR_1656|M.marinum_M --------------------------------------------------
MUL_1893|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4488|M.gilvum_PYR-GCK DHGAFPVPPLPVK-----------EPADA----
Mvan_1878|M.vanbaalenii_PYR-1 GDGAFPVPPLPVKTR-------ALEDADA----
MSMEG_2056|M.smegmatis_MC2_155 --GAYPVPPLPSVG--------TKETADA----
TH_1086|M.thermoresistible__bu HSGAFPVPSLPAGQQ-------RTENAKETVDV
MAB_2141|M.abscessus_ATCC_1997 MAGGFPVPPLPGQRVDTGAHPSTKEDADA----
Rv0084|M.tuberculosis_H37Rv ---------------------------------
Mb0087|M.bovis_AF2122/97 ---------------------------------
MAV_5112|M.avium_104 ---------------------------------
MMAR_1656|M.marinum_M ---------------------------------
MUL_1893|M.ulcerans_Agy99 ---------------------------------