For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YSTEALNHVISVAAASADQVDAEGAYPAAAVDALRGSGLLGLVVPEEAGGLGAGPQQFVDVVGGLAAACG STAMVYLMHTAAAVTVAAAPPPGMPEMLSAMASGNALGTLAFSEKGSRSHFWAPVSTAALSDDGDTVKLR ADKSWVTSAGYADIYIVSAGSTSGVAGEVDLYALPRSGEGLSVVAPFAGMGLRGNASSPMTVDATIPVAS RLGAQAAGFGLLMETVLPWFNLGNAAVSLGLAGSAYVATIGHVSGARLEHRDEALSALPTIRAQIAKMGT TLAAQRAYLRSAAASVSAPDEETLNHVLGIKASANDAALTITESAMRVCGGAAFSKHLPIERNFRDARAG AVMAPTADVLYDFYGKAVTGLPLF*
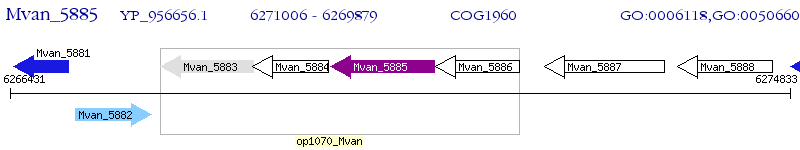
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5885 | - | - | 100% (375) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1476 | - | 7e-24 | 27.14% (350) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1719 | - | 1e-21 | 24.64% (349) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1706 | fadE16 | 1e-148 | 70.13% (375) | acyl-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4943 | - | 4e-22 | 26.29% (350) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv1679 | fadE16 | 1e-148 | 70.13% (375) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 7e-22 | 24.93% (349) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 1e-22 | 25.21% (349) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_1262 | fadE25 | 2e-21 | 24.36% (349) | acyl-CoA dehydrogenase FadE25 |
| M. avium 104 | MAV_2668 | - | 4e-22 | 25.65% (382) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3982 | - | 1e-24 | 28.57% (336) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4603 | - | 5e-22 | 27.30% (348) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_2626 | fadE25 | 4e-22 | 24.64% (349) | acyl-CoA dehydrogenase FadE25 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1262|M.marinum_M MAGWAGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADVDENSRFPQE
MUL_2626|M.ulcerans_Agy99 MAGWAGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADVDENSRFPQE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
MAB_3618c|M.abscessus_ATCC_199 ---MAGNPSFDLFKLGEEHDELRAAIRGLAEKEIAPHAKDVDEKARFPQE
MSMEG_3982|M.smegmatis_MC2_155 --------------MNTELLDFRAMMQDLASTQVAPYARDVDEQERFPEE
Mflv_4943|M.gilvum_PYR-GCK -------MPVDRLLPTDDARDLVELARQIADKVLDPIVDRHEKDETYPEG
MAV_2668|M.avium_104 -------MTVDRLLPTPEARELVRLAADVADKVLDPIVDEHEKNETYPEG
TH_4603|M.thermoresistible__bu -------MAVDRLLPSQEAAELIELTREIADKVLDPIVDRHEKDETYPEG
Mb1706|M.bovis_AF2122/97 --------------------MATPGVVQEVVSVAAEHAERVDTDCAFPAE
Rv1679|M.tuberculosis_H37Rv --------------------MATPGVVQEVVSVAAEHAERVDTDCAFPAE
Mvan_5885|M.vanbaalenii_PYR-1 --------------------MYSTEALNHVISVAAASADQVDAEGAYPAA
. . . : :*
MMAR_1262|M.marinum_M ALDALNASGFNAVHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MUL_2626|M.ulcerans_Agy99 ALDALNASGFNALHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MAB_3618c|M.abscessus_ATCC_199 ALDALVASGFNAVHVPEEYDGQGADSVAACIVIEEVARVCASSSLIPAVN
MSMEG_3982|M.smegmatis_MC2_155 AWQALCAADLPGLAHPTELGGSDGDLAAQAIAVEELSAACASTAMVLLVN
Mflv_4943|M.gilvum_PYR-GCK VFATLGEAGLLSLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVSVH
MAV_2668|M.avium_104 VFATLGETGLLTLPHPEQWGGGGQPYEVYLQVLEELAARWAAVAVAVSVH
TH_4603|M.thermoresistible__bu VFEQLGAAGLLSLPQPEEWGGGGQPYEVYLQVLEEIAARWASVAVAVSVH
Mb1706|M.bovis_AF2122/97 AVDALRKTGLLGLVLPREIGGMGSGPVEFTEVVAQLSAACGSTAMIYLMH
Rv1679|M.tuberculosis_H37Rv AVDALRKTGLLGLVLPREIGGMGSGPVEFTEVVAQLSAACGSTAMIYLMH
Mvan_5885|M.vanbaalenii_PYR-1 AVDALRGSGLLGLVVPEEAGGLGAGPQQFVDVVGGLAAACGSTAMVYLMH
. * :.: : * : .* . .: :: .: :: ::
MMAR_1262|M.marinum_M KLGTMGLILRGSDDLKKQVLPSLADGAAMASYALSEREAGSDAGAMRTRA
MUL_2626|M.ulcerans_Agy99 KLGTMGLILRGSDELKKQVLPSLADGAAMASYALSEREAGSDAGAMRTRA
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
MAB_3618c|M.abscessus_ATCC_199 KLGTMGLILNGSDELKKQVLPAIASGEAMASYALSEREAGSDAASMRTRA
MSMEG_3982|M.smegmatis_MC2_155 WAGTCTVIHQGSDELKKLVVPDVSTGAVGAGWCLTEPTGGSDLSGIRTKA
Mflv_4943|M.gilvum_PYR-GCK SLSCHPLMSFGSDEQRQRWLPDMLGGSTIGAYSLSEPQAGSDAAALTCRA
MAV_2668|M.avium_104 VLSCHPLIAFGTDEQQQEWLPTMLGGHTIGAYSLSEPQAGSDAAALTCKA
TH_4603|M.thermoresistible__bu SLSSHPLLVFGTEEQKKRWLPGMLSGEQIGAYSLSEPQAGSDAAALRCAA
Mb1706|M.bovis_AF2122/97 MAAAVTVAASPPPGLP-DLLADMASGKQLGTLAFSEPGSRSHFWAPVSTA
Rv1679|M.tuberculosis_H37Rv MAAAVTVAASPPPGLP-DLLADMASGKQLGTLAFSEPGSRSHFWAPVSTA
Mvan_5885|M.vanbaalenii_PYR-1 TAAAVTVAAAPPPGMP-EMLSAMASGNALGTLAFSEKGSRSHFWAPVSTA
. : . :. : . .::* . *. . *
MMAR_1262|M.marinum_M K--ADGDDWILNGTKCWITNGGKSSWYTVMAVTDPEKGANGISAFMVHKD
MUL_2626|M.ulcerans_Agy99 K--ADGDDWILNGTKCWITNGGKSSWYTVMAVTDPENGANGISAFVVHKD
MLBr_00737|M.leprae_Br4923 K--ADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHKD
MAB_3618c|M.abscessus_ATCC_199 K--ADGDDWIINGSKCWITNGGKSSWYTVMAVTDPEKKANGISAFMVHKD
MSMEG_3982|M.smegmatis_MC2_155 Q--RDGSDWVLNGQKRFISNADWGDWFAVLARSGEE---NEFGVFMVHKS
Mflv_4943|M.gilvum_PYR-GCK T--AVDGGYRITGSKAWITHGGIADFYNLFARTGEG--SQGISCFLVPRD
MAV_2668|M.avium_104 T--ATDGGYRVNGAKAWITHGGIADFYSLFARTGEG--GQGISCFLVDKD
TH_4603|M.thermoresistible__bu T--PTDGGYVINGSKSWITHGGKADFYTLFARTGEG--SRGVSCFLVPAD
Mb1706|M.bovis_AF2122/97 --SADGDGIAVRADKSWVTSAGFADVYVVSVGSADG-AAGDVDLYAVPAD
Rv1679|M.tuberculosis_H37Rv --SADGDGIAVRADKSWVTSAGFADVYVVSVGSADG-AAGDVDLYAVPAD
Mvan_5885|M.vanbaalenii_PYR-1 ALSDDGDTVKLRADKSWVTSAGYADIYIVSAGSTSG-VAGEVDLYALPRS
.. : . * ::: .. . : : . : .. : : .
MMAR_1262|M.marinum_M DEGFVVGPKERKLGIKGSPTTELYFENCRVPGDRMIGEPGTGFKTALATL
MUL_2626|M.ulcerans_Agy99 DEGFVVGPKERKLGIKGSPTTELYFENCRVPGDRMIGEPGTGFKTALATL
MLBr_00737|M.leprae_Br4923 DEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALATL
MAB_3618c|M.abscessus_ATCC_199 DEGFTVGPLEHKLGIKGSPTAELYFENCRIPGDRIIGEPGTGFKTALQTL
MSMEG_3982|M.smegmatis_MC2_155 DAGLSFGPPEKKMGLRGSPTADVILEDCRIPGERVVGDPTQGYKYIIDEL
Mflv_4943|M.gilvum_PYR-GCK TEGLTFGKPEEKMGLHAVPTTAAHYDDANLDSDRRIGTEGQGLQIAFSAL
MAV_2668|M.avium_104 TEGLTFGKPEEKMGLHAIPTTTAHYDNALVSAQRRIGAEGQGLQIAFSAL
TH_4603|M.thermoresistible__bu QPGLSFGKPEEKMGLHAVPTTSAFYDNARIDADRRIGEEGQGLQIAFSAL
Mb1706|M.bovis_AF2122/97 TPGLRVAGTFTGMGLRGNASAPMAVD-IRIPDSYRLGEAGGGFGIMMQTV
Rv1679|M.tuberculosis_H37Rv TPGLRVAGTFTGMGLRGNASAPMAVD-IRIPDSYRLGEAGGGFGIMMQTV
Mvan_5885|M.vanbaalenii_PYR-1 GEGLSVVAPFAGMGLRGNASSPMTVD-ATIPVASRLGAQAAGFGLLMETV
*: . :*::. .:: : : :* * : :
MMAR_1262|M.marinum_M D-HTRPTIGAQAVGIAQGALDAAIAYT--KDRKQFGKSISDFQAVQFMLA
MUL_2626|M.ulcerans_Agy99 D-HTRPTIGAQAVGIAQGALDAAIAYT--KDRKQFGKSISDFQAVQFMLA
MLBr_00737|M.leprae_Br4923 D-HTRPTIGAQAVGIAQGALDAAIVYT--KDRKQFGESISTFQSIQFMLA
MAB_3618c|M.abscessus_ATCC_199 D-HTRPTIGAQALGIAQGALDQAIAYT--KDRKQFGKSISDFQGVQFMLA
MSMEG_3982|M.smegmatis_MC2_155 N-GSRALIAAQALGLAKGALRTAIEYT--AERRQFGSALSTFQMVRAMVA
Mflv_4943|M.gilvum_PYR-GCK D-SGRLGIAAVAVGLAQAALDEAVAYS--QERTTFGRKIIDHQGLAFLLA
MAV_2668|M.avium_104 D-SGRLGIAAVAVGLAQAALDEAVSYA--QQRTTFGRKIIDHQGLAFLLA
TH_4603|M.thermoresistible__bu D-SGRLGIAAVATGLAQAALDEAVAYA--NERTAFGRKIIDHQGLGFLLA
Mb1706|M.bovis_AF2122/97 LPWFNLGNAAVSLGLATAATGAAVKHVGTARLEHLGGSLAELPTIRAQIA
Rv1679|M.tuberculosis_H37Rv LPWFNLGNAAVSLGLATAATGAAVKHVGTARLEHLGGSLAELPTIRAQIA
Mvan_5885|M.vanbaalenii_PYR-1 LPWFNLGNAAVSLGLAGSAYVATIGHVSGARLEHRDEALSALPTIRAQIA
. .* : *:* .* :: : . : : :*
MMAR_1262|M.marinum_M DMAMKVESARLMVYSAAARAERGESNLGFISAASKCLASDVAMEVTTDAV
MUL_2626|M.ulcerans_Agy99 DMAMKVESARLMVYSAAARAERGESNLGFISAASKCLASDVAMEVTTDAV
MLBr_00737|M.leprae_Br4923 DMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAV
MAB_3618c|M.abscessus_ATCC_199 DMAMKVEAARLMVYTAAARAERGEKNLGFISSASKCFASDVAMEVTTDAV
MSMEG_3982|M.smegmatis_MC2_155 DAAIRVESASALLYKAVDAIENKDPRARSLVSMAKVLCSDNAMAVTTDAV
Mflv_4943|M.gilvum_PYR-GCK DMAAAVDSARATYLDAARRRDAGLP-YSRHASVAKLVATDAAMKVTTDAV
MAV_2668|M.avium_104 DMAAAVDSARATYLDAARRRDAGLP-YSRNASVAKLIATDAAMKVTTDAV
TH_4603|M.thermoresistible__bu DMAAAVATARATYLDAARRRDQGRP-YSQQASIAKLTATDAAMKVTTDAV
Mb1706|M.bovis_AF2122/97 RMGTTLAAQKAYLEVAANSVSSPDDTTLTHVLGVKASVNDAALTITESAM
Rv1679|M.tuberculosis_H37Rv RMGTTLAAQKAYLEVAANSVSSPDDTTLTHVLGVKASVNDAALTITESAM
Mvan_5885|M.vanbaalenii_PYR-1 KMGTTLAAQRAYLRSAAASVSAPDEETLNHVLGIKASANDAALTITESAM
. : : *. . * .* *: :* .*:
MMAR_1262|M.marinum_M QLFGGAGYTVDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR-----
MUL_2626|M.ulcerans_Agy99 QLFGGAGYTVDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR-----
MLBr_00737|M.leprae_Br4923 QLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR-----
MAB_3618c|M.abscessus_ATCC_199 QLFGGAGYTTDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLTS----
MSMEG_3982|M.smegmatis_MC2_155 QLLGGYGFTREYPVERMMRDAKVTQIYEGSNQIQRLVISKGLYKGEALS
Mflv_4943|M.gilvum_PYR-GCK QVLGGAGYTRDYRVERYMREAKITQIFEGTNQIQRLVISRHLTRG----
MAV_2668|M.avium_104 QVFGGYGYTRDYRVERYMREAKITQIFEGTNQIQRLVIGRSLVGS----
TH_4603|M.thermoresistible__bu QVFGGVGYTRDYRVERYMREAKIMQIFEGTNQIQRLVIARGLTR-----
Mb1706|M.bovis_AF2122/97 RVCGGAAFSKHLPIERAFRDARAGSVMAPTADALYDFYGRAVTGLPLF-
Rv1679|M.tuberculosis_H37Rv RVCGGAAFSKHLPIERAFRDARAGSVMAPTADALYDFYGRAVTGLPLF-
Mvan_5885|M.vanbaalenii_PYR-1 RVCGGAAFSKHLPIERNFRDARAGAVMAPTADVLYDFYGKAVTGLPLF-
:: ** .:: . :** :*:*: : : : . .: :