For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VILDLMRLMATLADGAVGEIAVASVAERETAEKWCARSGNTVVQADLDETGAGTLTVRRGRPPDPLLVLG PERMPGARLWMYTNFHCNLACDYCCVESSPQAPRRELGADRIARLAREAADWGVREVFLTGGEPFLLSDI GTIVRSCAALLPTTVLTNGMVFRGRGLRELDAIPRDNVALQISLDSATSELHDSHRGHGSWARAVEGIRV ARGLGFRVRVAATVAAPVPGELKAFHDFLDELGIAQDDQVVRPVAREGSAADGVQITRASLVPEVAVTAE GVYWHPVAAIDDDALVTRQIEPLAPALDEIAHAFAELWARKANAAQLFPCA*
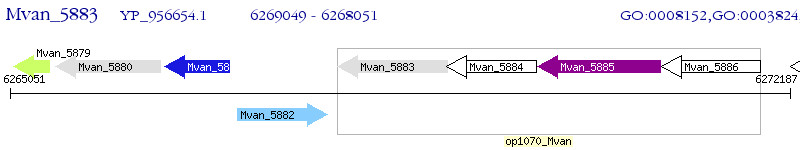
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5883 | - | - | 100% (332) | radical SAM domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1296 | - | 7e-12 | 30.70% (215) | radical SAM domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_5047 | moaA | 9e-07 | 31.17% (154) | molybdenum cofactor biosynthesis protein A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1708 | moeX | 1e-134 | 67.98% (331) | molybdopterin biosynthesis protein MoeX |
| M. gilvum PYR-GCK | Mflv_5060 | - | 2e-11 | 30.48% (210) | radical SAM domain-containing protein |
| M. tuberculosis H37Rv | Rv1681 | moeX | 1e-134 | 67.98% (331) | molybdopterin biosynthesis protein MoeX |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3835c | - | 2e-10 | 28.57% (196) | coenzyme PQQ synthesis protein E PqqE |
| M. marinum M | MMAR_1021 | pqqE | 5e-11 | 30.26% (195) | coenzyme PQQ synthesis protein E, PqqE |
| M. avium 104 | MAV_4478 | - | 3e-10 | 30.05% (193) | radical SAM domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3721 | pqqE | 1e-10 | 33.08% (133) | pyrroloquinoline quinone biosynthesis protein PqqE |
| M. thermoresistible (build 8) | TH_1901 | pqqE | 5e-12 | 31.05% (190) | PROBABLE COENZYME PQQ SYNTHESIS PROTEIN E PQQE (COENZYME PQQ |
| M. ulcerans Agy99 | MUL_0773 | pqqE | 4e-11 | 30.26% (195) | coenzyme PQQ synthesis protein E, PqqE |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1021|M.marinum_M ---------MTTA--VPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
MUL_0773|M.ulcerans_Agy99 ---------MTTA--VPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
MAV_4478|M.avium_104 ---------MTTAAPVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
TH_1901|M.thermoresistible__bu ---------------VPRLIEQFEQGLDAPICLTWELTYACNLACVHCLS
Mflv_5060|M.gilvum_PYR-GCK ---------MTLAAPVPRLVDQFERGLDAPICLTWELTYACNLSCVHCLS
MAB_3835c|M.abscessus_ATCC_199 MSAPAVPRLNVAAVAAPKLVDQFEQGLDAPICLTWELTYACNLSCVHCLS
Mb1708|M.bovis_AF2122/97 -----------MIIELMRRVVGLAQGATAEVAVYGDR-------------
Rv1681|M.tuberculosis_H37Rv -----------MIIELMRRVVGLAQGATAEVAVYGDR-------------
Mvan_5883|M.vanbaalenii_PYR-1 ----------MVILDLMRLMATLADGAVGEIAVASVA-------------
MSMEG_3721|M.smegmatis_MC2_155 ----------MDRPYALLAELTYACPLHCPYCSNPVALQN----------
.
MMAR_1021|M.marinum_M SSGKRDPGELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
MUL_0773|M.ulcerans_Agy99 SSGKRDPGELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
MAV_4478|M.avium_104 SSGKRDPRELSTRQCMDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
TH_1901|M.thermoresistible__bu SSGKRDPRELSTQQCKDIIDELQRMQVFYVNIGGGEPTVRSDFWELVDYA
Mflv_5060|M.gilvum_PYR-GCK SSGKRDPRELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
MAB_3835c|M.abscessus_ATCC_199 SSGKRDPRELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
Mb1708|M.bovis_AF2122/97 ------DRDLAERWCANTGNTLVRADVDQT--GVGTLVVR----------
Rv1681|M.tuberculosis_H37Rv ------DRDLAERWCANTGNTLVRADVDQT--GVGTLVVR----------
Mvan_5883|M.vanbaalenii_PYR-1 ------ERETAEKWCARSGNTVVQADLDET--GAGTLTVR----------
MSMEG_3721|M.smegmatis_MC2_155 -----YRDELSTVEWQRVFAEAAELGVLQVHLSGGEPMQRHDIVDLVRCA
: : . : . . * *
MMAR_1021|M.marinum_M TAHHVGVKFSTNGVRINPEVAARLAASDYVDVQISLDGATAEVNDAVRGA
MUL_0773|M.ulcerans_Agy99 TAHHVGVKFSTNGVRINPEVAARLAASDCVDVQISLDGATAEVNDAVRGA
MAV_4478|M.avium_104 TAHHVGVKFSTNGVRITPEVAARLAASDYVDVQISLDGATAEVNDAVRGP
TH_1901|M.thermoresistible__bu TAHNVGVKFSTNGVRITPEVARRLAASDYVDVQISLDGATAEINDAVRGA
Mflv_5060|M.gilvum_PYR-GCK TEHHVGVKFSTNGVRIDKTVAAKLAASDYVDVQISLDGATAEVNDAVRGP
MAB_3835c|M.abscessus_ATCC_199 TEHHVGVKFSTNGVRITEEVAARLAASDYVDVQISLDGATAEVNDAVRGA
Mb1708|M.bovis_AF2122/97 -RGHPPDPASVLGPDRLPGVRLWLYTNFHCNLCCDYCCVSSSPSTPHREL
Rv1681|M.tuberculosis_H37Rv -RGHPPDPASVLGPDRLPGVRLWLYTNFHCNLCCDYCCVSSSPSTPHREL
Mvan_5883|M.vanbaalenii_PYR-1 -RGRPPDPLLVLGPERMPGARLWMYTNFHCNLACDYCCVESSPQAPRREL
MSMEG_3721|M.smegmatis_MC2_155 NGLGMYTNLITSGLGFSTRRAEQLRDAGLDHVQVSIQADQPEVSDRIAGT
. * : .: . .. .
MMAR_1021|M.marinum_M GSFAMAVRALENLAAAGFAD---AKISVVVTRHNVGQLDDFAALADRYGA
MUL_0773|M.ulcerans_Agy99 GSFAMAVRALENLAAAGFAD---AKISVVVTRHNVGQLDDFAALADRYGA
MAV_4478|M.avium_104 GSFDMAVRALQNLADAGFTQNGGAKISVVVTRHNVGQLDEFAALASRYGA
TH_1901|M.thermoresistible__bu GSFDMAVRALQNLADAGFAD---AKISVVVTRHNVDQLDDFKALADRYGA
Mflv_5060|M.gilvum_PYR-GCK GSFAMAVRALENLKEAGFKD---AKISVVVTRHNIDQLDDFKALADTYGA
MAB_3835c|M.abscessus_ATCC_199 GSFDMAVRALENLAAAGFKD---AKISVVVTRHNVDQLDEFASLAARYGA
Mb1708|M.bovis_AF2122/97 G-AERIGRIVGEAARWGVRE-------LFLTGGEPFLLPDIDTIIATCVK
Rv1681|M.tuberculosis_H37Rv G-AERIGRIVGEAARWGVRE-------LFLTGGEPFLLPDIDTIIATCVK
Mvan_5883|M.vanbaalenii_PYR-1 G-ADRIARLAREAADWGVRE-------VFLTGGEPFLLSDIGTIVRSCAA
MSMEG_3721|M.smegmatis_MC2_155 ASFERKHDVARLVKKLGWPLT----LNVVLHRQNIDRIGAILDMAAVMEA
. * :.: : : : :
MMAR_1021|M.marinum_M TLRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFFHL
MUL_0773|M.ulcerans_Agy99 TLRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFFHL
MAV_4478|M.avium_104 TLRITRLRPSGRGADVWDELHPTADQQVQLYDWLVAKGERVLTGDSFFHL
TH_1901|M.thermoresistible__bu TLRITRLRPSGRGADVWDELHPTAEQQVQLYNWLVAHGEKVLTGDSFFHL
Mflv_5060|M.gilvum_PYR-GCK TLRITRLRPSGRGADVWDDLHPTADQQVQLYNWLVAHGERVLTGDSFFHL
MAB_3835c|M.abscessus_ATCC_199 TLRITRLRPSGRGADVWDELHPTAAQQRQLYDWLVRNGERVLTGDSFFHL
Mb1708|M.bovis_AF2122/97 QLPTTVLTNG----MVFKGRGRRALESLPRG-LALQISLDSATPELHDAH
Rv1681|M.tuberculosis_H37Rv QLPTTVLTNG----MVFKGRGRRALESLPRG-LALQISLDSATPELHDAH
Mvan_5883|M.vanbaalenii_PYR-1 LLPTTVLTNG----MVFRGRGLRELDAIPRDNVALQISLDSATSELHDSH
MSMEG_3721|M.smegmatis_MC2_155 DRIELANTQYYGWAGRNRAELLPSRDQLERAQTVVR-AARGRLGNRMEII
: : . :
MMAR_1021|M.marinum_M APLGS-SGALAGLNMCG-AGRVVCLIDPVGDVYACPFAIHDR---FLAGN
MUL_0773|M.ulcerans_Agy99 APLGS-SGALAGLNMCG-AGRVVCLIDPVGDVYACPFAIHDR---FLAGN
MAV_4478|M.avium_104 APLGQ-SGALAGLNMCG-AGRVVCLIDPVGDVYACPFAIHDR---FLAGN
TH_1901|M.thermoresistible__bu SGLGE-PGALAGLNLCG-AGRVVCLIDPVGDVYACPFAIHDR---FLAGN
Mflv_5060|M.gilvum_PYR-GCK SGLGE-PGALAGLNLCG-AGRVVCLIDPVGDVYACPFAIHDK---FLAGN
MAB_3835c|M.abscessus_ATCC_199 SAYGDGSGGLPGLNMCG-AGRVVCLIDPVGDVYACPFAIHDR---FLAGN
Mb1708|M.bovis_AF2122/97 RGAGTWVKAVAGIRLALSLGFRVRVAATVASPAPGELTAFHD---FLDGL
Rv1681|M.tuberculosis_H37Rv RGAGTWVKAVAGIRLALSLGFRVRVAATVASPAPGELTAFHD---FLDGL
Mvan_5883|M.vanbaalenii_PYR-1 RGHGSWARAVEGIRVARGLGFRVRVAATVAAPVPGELKAFHD---FLDEL
MSMEG_3721|M.smegmatis_MC2_155 YVIPDYYEKYPKPCMAG-WARRQFTVVPNGEALPCPAAHELARGLSLPGT
:. . . . . *
MMAR_1021|M.marinum_M VLTDGGFDQVWKNAPLFRQLR-EPQSAGACGSCGHYDSCRGGCMAAKFFT
MUL_0773|M.ulcerans_Agy99 VLTDGGFDQVWKNAPLFRQLR-EPQSAGACGSCGHYDSCRGGCMAAKFFT
MAV_4478|M.avium_104 VLSDGGFDNVWKNAPLFRELR-EPQSAGACGSCGHYDSCRGGCMAAKFFT
TH_1901|M.thermoresistible__bu VLTDGGFDNVWKNSQLFRELR-EPQSAGACSGCSHFDSCRGGCMAAKFFT
Mflv_5060|M.gilvum_PYR-GCK ILTSNGFEDVWQNSELFRELR-EPQSAGACSGCNHYDACRGGCMAAKFFT
MAB_3835c|M.abscessus_ATCC_199 VLSDNGFDQVWKHSQLFSELR-EPQSAGACGSCGHYDSCRGGCMAAKFFT
Mb1708|M.bovis_AF2122/97 GIAPG--------DQLVRPIA-LEGAASQGVALTRESLVPEVTVTADGVY
Rv1681|M.tuberculosis_H37Rv GIAPG--------DQLVRPIA-LEGAASQGVALTRESLVPEVTVTADGVY
Mvan_5883|M.vanbaalenii_PYR-1 GIAQD--------DQVVRPVA-REGSAADGVQITRASLVPEVAVTAEGVY
MSMEG_3721|M.smegmatis_MC2_155 NVRRHSLEWIWMQSPLFRQFRGDDWMPDPCRTCDRREIDFGGCRCQAFAL
: :. . . : .
MMAR_1021|M.marinum_M GLPLDGPDPECVQG----YGAPALAQERHAPRPRVDHSRGSRV----SAP
MUL_0773|M.ulcerans_Agy99 GLPLDGPDPECVQG----YGAPALAQERHAPRPRVDHSRGSRE-------
MAV_4478|M.avium_104 GLPLDGPDPECVQG----HSAAALAGDREAPRPRADHSRGKRI----RQP
TH_1901|M.thermoresistible__bu GLPLDGPDPECVEG----HGAPALARERTKPKPRVDHSRGG--------P
Mflv_5060|M.gilvum_PYR-GCK GLPMDGPDPECVQG----YGEPALAADRDKPKSSVDHSRSGGRG-KPKGP
MAB_3835c|M.abscessus_ATCC_199 GLPMDGPDPECVQG----YGEAALAADRVVPKPSGDHSHSKSKRGPATGP
Mb1708|M.bovis_AF2122/97 WHPVAATDERALVT----RTVEPLTPALDMVSRLFAEQWTRAAEEAALFP
Rv1681|M.tuberculosis_H37Rv WHPVAATDERALVT----RTVEPLTPALDMVSRLFAEQWTRAAEEAALFP
Mvan_5883|M.vanbaalenii_PYR-1 WHPVAAIDDDALVT----RQIEPLAPALDEIAHAFAELWARKANAAQLFP
MSMEG_3721|M.smegmatis_MC2_155 TGDAARTDPVCHLSPDHGLVETAVAAANDPTLPHDGVMIPRPNPGRTRTQ
* . .::
MMAR_1021|M.marinum_M VPLTLTTR--------PPARLCNESPV-------
MUL_0773|M.ulcerans_Agy99 ----------------------------------
MAV_4478|M.avium_104 VPLTLSVRPPVLTPAAPPARLCNESPV-------
TH_1901|M.thermoresistible__bu VALKLLTK--------PPARLCNESPV-------
Mflv_5060|M.gilvum_PYR-GCK IPLTLLSA--------PPKKFCNESPV-------
MAB_3835c|M.abscessus_ATCC_199 VALTLTRR--------PPSRACDENPLSTLRAGR
Mb1708|M.bovis_AF2122/97 CA--------------------------------
Rv1681|M.tuberculosis_H37Rv CA--------------------------------
Mvan_5883|M.vanbaalenii_PYR-1 CA--------------------------------
MSMEG_3721|M.smegmatis_MC2_155 LPLEVRR---------------------------