For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTAVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLSSSGKRDPGELSTRQCQDIIDELERMQVFYVN IGGGEPTVRPDFWELVDYATAHHVGVKFSTNGVRINPEVAARLAASDCVDVQISLDGATAEVNDAVRGAG SFAMAVRALENLAAAGFADAKISVVVTRHNVGQLDDFAALADRYGATLRITRLRPSGRGADVWEELHPTA AQQVALYDWLVAKGERVLTGDSFFHLAPLGSSGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDRFLAG NVLTDGGFDQVWKNAPLFRQLREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLDGPDPECVQGYGAPAL AQERHAPRPRVDHSRGSRE
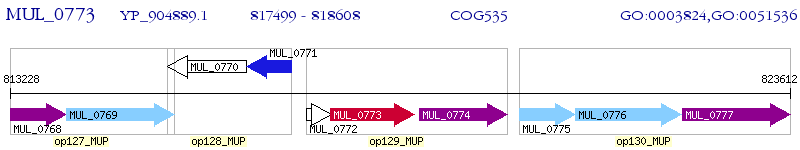
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0773 | pqqE | - | 100% (369) | coenzyme PQQ synthesis protein E, PqqE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0712 | pqqE | 0.0 | 88.86% (368) | coenzyme PQQ synthesis protein E |
| M. gilvum PYR-GCK | Mflv_5060 | - | 0.0 | 83.56% (365) | radical SAM domain-containing protein |
| M. tuberculosis H37Rv | Rv0693 | pqqE | 0.0 | 88.86% (368) | coenzyme PQQ synthesis protein E |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3835c | - | 0.0 | 86.46% (362) | coenzyme PQQ synthesis protein E PqqE |
| M. marinum M | MMAR_1021 | pqqE | 0.0 | 99.73% (368) | coenzyme PQQ synthesis protein E, PqqE |
| M. avium 104 | MAV_4478 | - | 0.0 | 91.42% (373) | radical SAM domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_1423 | - | 0.0 | 84.93% (365) | radical SAM domain-containing protein |
| M. thermoresistible (build 8) | TH_1901 | pqqE | 0.0 | 88.67% (362) | PROBABLE COENZYME PQQ SYNTHESIS PROTEIN E PQQE (COENZYME PQQ |
| M. vanbaalenii PYR-1 | Mvan_1296 | - | 0.0 | 86.30% (365) | radical SAM domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1423|M.smegmatis_MC2_155 ---------MTSVQPVPRLVEQFERGLDAPICLTWELTYACNLACVHCLS
TH_1901|M.thermoresistible__bu ---------------VPRLIEQFEQGLDAPICLTWELTYACNLACVHCLS
Mflv_5060|M.gilvum_PYR-GCK ---------MTLAAPVPRLVDQFERGLDAPICLTWELTYACNLSCVHCLS
Mvan_1296|M.vanbaalenii_PYR-1 ---------MTLAAPVPRLVDQFERGLDAPICLTWELTYACNLSCVHCLS
MAB_3835c|M.abscessus_ATCC_199 MSAPAVPRLNVAAVAAPKLVDQFEQGLDAPICLTWELTYACNLSCVHCLS
MUL_0773|M.ulcerans_Agy99 -----------MTTAVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
MMAR_1021|M.marinum_M -----------MTTAVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
Mb0712|M.bovis_AF2122/97 -----------MTSPVPRLIEQFERGLDAPICLTWELTYACNLACVHCLS
Rv0693|M.tuberculosis_H37Rv -----------MTSPVPRLIEQFERGLDAPICLTWELTYACNLACVHCLS
MAV_4478|M.avium_104 ---------MTTAAPVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
.*:*::***:******************:******
MSMEG_1423|M.smegmatis_MC2_155 SSGKRDPRELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
TH_1901|M.thermoresistible__bu SSGKRDPRELSTQQCKDIIDELQRMQVFYVNIGGGEPTVRSDFWELVDYA
Mflv_5060|M.gilvum_PYR-GCK SSGKRDPRELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
Mvan_1296|M.vanbaalenii_PYR-1 SSGKRDPRELSTRQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
MAB_3835c|M.abscessus_ATCC_199 SSGKRDPRELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDYA
MUL_0773|M.ulcerans_Agy99 SSGKRDPGELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
MMAR_1021|M.marinum_M SSGKRDPGELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
Mb0712|M.bovis_AF2122/97 SSGKRDPGELSTRQCKDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
Rv0693|M.tuberculosis_H37Rv SSGKRDPGELSTRQCKDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
MAV_4478|M.avium_104 SSGKRDPRELSTRQCMDIIDELERMQVFYVNIGGGEPTVRPDFWELVDYA
******* ****:** ******:*****************.*********
MSMEG_1423|M.smegmatis_MC2_155 TAHHVGVKFSTNGVRITPEVAAKLAASDYVDVQISLDGANAEVNDAVRGK
TH_1901|M.thermoresistible__bu TAHNVGVKFSTNGVRITPEVARRLAASDYVDVQISLDGATAEINDAVRGA
Mflv_5060|M.gilvum_PYR-GCK TEHHVGVKFSTNGVRIDKTVAAKLAASDYVDVQISLDGATAEVNDAVRGP
Mvan_1296|M.vanbaalenii_PYR-1 TEHHVGVKFSTNGVRITPEVAAKLAASDYVDVQISLDGATAEVNDTVRGP
MAB_3835c|M.abscessus_ATCC_199 TEHHVGVKFSTNGVRITEEVAARLAASDYVDVQISLDGATAEVNDAVRGA
MUL_0773|M.ulcerans_Agy99 TAHHVGVKFSTNGVRINPEVAARLAASDCVDVQISLDGATAEVNDAVRGA
MMAR_1021|M.marinum_M TAHHVGVKFSTNGVRINPEVAARLAASDYVDVQISLDGATAEVNDAVRGA
Mb0712|M.bovis_AF2122/97 TAHHVGVKFSTNGVRITPEVATRLAATDYVDVQISLDGATAEVNDAIRGT
Rv0693|M.tuberculosis_H37Rv TAHHVGVKFSTNGVRITPEVATRLAATDYVDVQISLDGATAEVNDAIRGT
MAV_4478|M.avium_104 TAHHVGVKFSTNGVRITPEVAARLAASDYVDVQISLDGATAEVNDAVRGP
* *:************ ** :***:* **********.**:**::**
MSMEG_1423|M.smegmatis_MC2_155 GSFDMAVRALENLSNAGFT---DAKISVVVTRQNVDQLDEFAALAARYGA
TH_1901|M.thermoresistible__bu GSFDMAVRALQNLADAGFA---DAKISVVVTRHNVDQLDDFKALADRYGA
Mflv_5060|M.gilvum_PYR-GCK GSFAMAVRALENLKEAGFK---DAKISVVVTRHNIDQLDDFKALADTYGA
Mvan_1296|M.vanbaalenii_PYR-1 GSFAMAVRALENLKAAGFK---DAKISVVVTRHNIDQLDDFKALADTYGA
MAB_3835c|M.abscessus_ATCC_199 GSFDMAVRALENLAAAGFK---DAKISVVVTRHNVDQLDEFASLAARYGA
MUL_0773|M.ulcerans_Agy99 GSFAMAVRALENLAAAGFA---DAKISVVVTRHNVGQLDDFAALADRYGA
MMAR_1021|M.marinum_M GSFAMAVRALENLAAAGFA---DAKISVVVTRHNVGQLDDFAALADRYGA
Mb0712|M.bovis_AF2122/97 GSFDMAVRALQNLAAAGFA---GVKISVVITRRNVAQLDEFATLASRYGA
Rv0693|M.tuberculosis_H37Rv GSFDMAVRALQNLAAAGFA---GVKISVVITRRNVAQLDEFATLASRYGA
MAV_4478|M.avium_104 GSFDMAVRALQNLADAGFTQNGGAKISVVVTRHNVGQLDEFAALASRYGA
*** ******:** *** ..*****:**:*: ***:* :** ***
MSMEG_1423|M.smegmatis_MC2_155 TLRITRLRPSGRGADVWDDLHPTAEQQRQLYDWLVAKGDRVLTGDSFFHL
TH_1901|M.thermoresistible__bu TLRITRLRPSGRGADVWDELHPTAEQQVQLYNWLVAHGEKVLTGDSFFHL
Mflv_5060|M.gilvum_PYR-GCK TLRITRLRPSGRGADVWDDLHPTADQQVQLYNWLVAHGERVLTGDSFFHL
Mvan_1296|M.vanbaalenii_PYR-1 TLRITRLRPSGRGADVWDELHPTPAQQVQLYDWLVAHGERVLTGDSFFHL
MAB_3835c|M.abscessus_ATCC_199 TLRITRLRPSGRGADVWDELHPTAAQQRQLYDWLVRNGERVLTGDSFFHL
MUL_0773|M.ulcerans_Agy99 TLRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFFHL
MMAR_1021|M.marinum_M TLRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFFHL
Mb0712|M.bovis_AF2122/97 TLRITRLRPSGRGTDVWADLHPTADQQVQLYDWLVSKGERVLTGDSFFHL
Rv0693|M.tuberculosis_H37Rv TLRITRLRPSGRGTDVWADLHPTADQQVQLYDWLVSKGERVLTGDSFFHL
MAV_4478|M.avium_104 TLRITRLRPSGRGADVWDELHPTADQQVQLYDWLVAKGERVLTGDSFFHL
*************:*** :****. ** **:*** :*::**********
MSMEG_1423|M.smegmatis_MC2_155 SGLGA-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDKFLAGNILSD
TH_1901|M.thermoresistible__bu SGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDRFLAGNVLTD
Mflv_5060|M.gilvum_PYR-GCK SGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDKFLAGNILTS
Mvan_1296|M.vanbaalenii_PYR-1 SGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDKFLAGNVLSD
MAB_3835c|M.abscessus_ATCC_199 SAYGDGSGGLPGLNMCGAGRVVCLIDPVGDVYACPFAIHDRFLAGNVLSD
MUL_0773|M.ulcerans_Agy99 APLGS-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDRFLAGNVLTD
MMAR_1021|M.marinum_M APLGS-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDRFLAGNVLTD
Mb0712|M.bovis_AF2122/97 APLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDHFLAGNVLSD
Rv0693|M.tuberculosis_H37Rv APLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDHFLAGNVLSD
MAV_4478|M.avium_104 APLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDRFLAGNVLSD
: * .*.*.***:*************************:*****:*:.
MSMEG_1423|M.smegmatis_MC2_155 GGFQNVWQHSELFRELREPQSAGACASCGHFDACRGGCMAAKFFTGLPLD
TH_1901|M.thermoresistible__bu GGFDNVWKNSQLFRELREPQSAGACSGCSHFDSCRGGCMAAKFFTGLPLD
Mflv_5060|M.gilvum_PYR-GCK NGFEDVWQNSELFRELREPQSAGACSGCNHYDACRGGCMAAKFFTGLPMD
Mvan_1296|M.vanbaalenii_PYR-1 GGFDTVWKHSELFRELREPQSAGACSGCGHYDACRGGCMAAKFFTGLPMD
MAB_3835c|M.abscessus_ATCC_199 NGFDQVWKHSQLFSELREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPMD
MUL_0773|M.ulcerans_Agy99 GGFDQVWKNAPLFRQLREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLD
MMAR_1021|M.marinum_M GGFDQVWKNAPLFRQLREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLD
Mb0712|M.bovis_AF2122/97 GGFQNVWKNSSLFRELREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLD
Rv0693|M.tuberculosis_H37Rv GGFQNVWKNSSLFRELREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLD
MAV_4478|M.avium_104 GGFDNVWKNAPLFRELREPQSAGACGSCGHYDSCRGGCMAAKFFTGLPLD
.**: **::: ** :**********..*.*:*:***************:*
MSMEG_1423|M.smegmatis_MC2_155 GPDPECVEGWGAPALEKERVKPKPSGDHSRGTKQ-----GPVALKLLTKP
TH_1901|M.thermoresistible__bu GPDPECVEGHGAPALARERTKPKPRVDHSRG--------GPVALKLLTKP
Mflv_5060|M.gilvum_PYR-GCK GPDPECVQGYGEPALAADRDKPKSSVDHSRSGGRG-KPKGPIPLTLLSAP
Mvan_1296|M.vanbaalenii_PYR-1 GPDPECVQGYGEPALALDRDKPRSSVDHSRSGGR--TPKGPIPLTLLSAP
MAB_3835c|M.abscessus_ATCC_199 GPDPECVQGYGEAALAADRVVPKPSGDHSHSKSKRGPATGPVALTLTRRP
MUL_0773|M.ulcerans_Agy99 GPDPECVQGYGAPALAQERHAPRPRVDHSRGSRE----------------
MMAR_1021|M.marinum_M GPDPECVQGYGAPALAQERHAPRPRVDHSRGSRVS----APVPLTLTTRP
Mb0712|M.bovis_AF2122/97 GPDPECVQGHSEPALARERHLPRPRADHSRGRRVS----KPVPLTLSMRP
Rv0693|M.tuberculosis_H37Rv GPDPECVQGHSEPALARERHLPRPRADHSRGRRVS----KPVPLTLSMRP
MAV_4478|M.avium_104 GPDPECVQGHSAAALAGDREAPRPRADHSRGKRIR----QPVPLTLSVRP
*******:* . .** :* *:. ***:.
MSMEG_1423|M.smegmatis_MC2_155 --------PARFCNESPV-------
TH_1901|M.thermoresistible__bu --------PARLCNESPV-------
Mflv_5060|M.gilvum_PYR-GCK --------PKKFCNESPV-------
Mvan_1296|M.vanbaalenii_PYR-1 --------PKKFCNESPV-------
MAB_3835c|M.abscessus_ATCC_199 --------PSRACDENPLSTLRAGR
MUL_0773|M.ulcerans_Agy99 -------------------------
MMAR_1021|M.marinum_M --------PARLCNESPV-------
Mb0712|M.bovis_AF2122/97 --------PKRPCNESPV-------
Rv0693|M.tuberculosis_H37Rv --------PKRPCNESPV-------
MAV_4478|M.avium_104 PVLTPAAPPARLCNESPV-------