For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DRPYALLAELTYACPLHCPYCSNPVALQNYRDELSTVEWQRVFAEAAELGVLQVHLSGGEPMQRHDIVDL VRCANGLGMYTNLITSGLGFSTRRAEQLRDAGLDHVQVSIQADQPEVSDRIAGTASFERKHDVARLVKKL GWPLTLNVVLHRQNIDRIGAILDMAAVMEADRIELANTQYYGWAGRNRAELLPSRDQLERAQTVVRAARG RLGNRMEIIYVIPDYYEKYPKPCMAGWARRQFTVVPNGEALPCPAAHELARGLSLPGTNVRRHSLEWIWM QSPLFRQFRGDDWMPDPCRTCDRREIDFGGCRCQAFALTGDAARTDPVCHLSPDHGLVETAVAAANDPTL PHDGVMIPRPNPGRTRTQLPLEVRR*
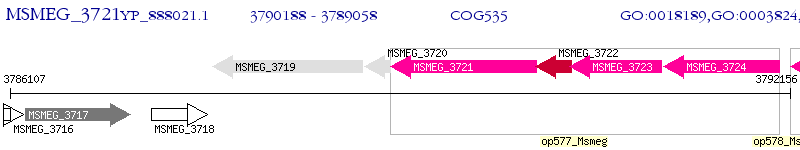
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3721 | pqqE | - | 100% (376) | pyrroloquinoline quinone biosynthesis protein PqqE |
| M. smegmatis MC2 155 | MSMEG_1423 | - | 6e-18 | 24.78% (335) | radical SAM domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5698 | moaA | 1e-07 | 35.71% (112) | molybdenum cofactor biosynthesis protein A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0712 | pqqE | 2e-18 | 25.13% (374) | coenzyme PQQ synthesis protein E |
| M. gilvum PYR-GCK | Mflv_5060 | - | 2e-18 | 25.93% (351) | radical SAM domain-containing protein |
| M. tuberculosis H37Rv | Rv0693 | pqqE | 2e-18 | 25.13% (374) | coenzyme PQQ synthesis protein E |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3835c | - | 2e-19 | 25.86% (348) | coenzyme PQQ synthesis protein E PqqE |
| M. marinum M | MMAR_1021 | pqqE | 2e-17 | 26.63% (338) | coenzyme PQQ synthesis protein E, PqqE |
| M. avium 104 | MAV_4478 | - | 5e-18 | 24.74% (380) | radical SAM domain-containing protein |
| M. thermoresistible (build 8) | TH_1901 | pqqE | 6e-17 | 24.86% (370) | PROBABLE COENZYME PQQ SYNTHESIS PROTEIN E PQQE (COENZYME PQQ |
| M. ulcerans Agy99 | MUL_0773 | pqqE | 2e-17 | 26.63% (338) | coenzyme PQQ synthesis protein E, PqqE |
| M. vanbaalenii PYR-1 | Mvan_1296 | - | 5e-16 | 25.23% (333) | radical SAM domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0712|M.bovis_AF2122/97 -----------MTSPVPRLIEQFERGLDAPICLTWELTYACNLACVHCLS
Rv0693|M.tuberculosis_H37Rv -----------MTSPVPRLIEQFERGLDAPICLTWELTYACNLACVHCLS
MAV_4478|M.avium_104 ---------MTTAAPVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
MMAR_1021|M.marinum_M -----------MTTAVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
MUL_0773|M.ulcerans_Agy99 -----------MTTAVPRLIEQFEHGLDAPICLTWELTYACNLACVHCLS
Mflv_5060|M.gilvum_PYR-GCK ---------MTLAAPVPRLVDQFERGLDAPICLTWELTYACNLSCVHCLS
Mvan_1296|M.vanbaalenii_PYR-1 ---------MTLAAPVPRLVDQFERGLDAPICLTWELTYACNLSCVHCLS
TH_1901|M.thermoresistible__bu ---------------VPRLIEQFEQGLDAPICLTWELTYACNLACVHCLS
MAB_3835c|M.abscessus_ATCC_199 MSAPAVPRLNVAAVAAPKLVDQFEQGLDAPICLTWELTYACNLSCVHCLS
MSMEG_3721|M.smegmatis_MC2_155 --------------------------MDRPYALLAELTYACPLHCPYCSN
:* * .* ****** * * :* .
Mb0712|M.bovis_AF2122/97 SSGKRDPG-ELSTRQCKDIIDELERMQVFYVNIGGGEPTVRPDFWELVDY
Rv0693|M.tuberculosis_H37Rv SSGKRDPG-ELSTRQCKDIIDELERMQVFYVNIGGGEPTVRPDFWELVDY
MAV_4478|M.avium_104 SSGKRDPR-ELSTRQCMDIIDELERMQVFYVNIGGGEPTVRPDFWELVDY
MMAR_1021|M.marinum_M SSGKRDPG-ELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDY
MUL_0773|M.ulcerans_Agy99 SSGKRDPG-ELSTRQCQDIIDELERMQVFYVNIGGGEPTVRPDFWELVDY
Mflv_5060|M.gilvum_PYR-GCK SSGKRDPR-ELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDY
Mvan_1296|M.vanbaalenii_PYR-1 SSGKRDPR-ELSTRQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDY
TH_1901|M.thermoresistible__bu SSGKRDPR-ELSTQQCKDIIDELQRMQVFYVNIGGGEPTVRSDFWELVDY
MAB_3835c|M.abscessus_ATCC_199 SSGKRDPR-ELSTQQCKDIIDELERMQVFYVNIGGGEPTVRSDFWELVDY
MSMEG_3721|M.smegmatis_MC2_155 PVALQNYRDELSTVEWQRVFAEAAELGVLQVHLSGGEPMQRHDIVDLVRC
. . :: **** : :: * .: *: *::.**** * *: :**
Mb0712|M.bovis_AF2122/97 ATAHHVGVKFSTNGVRITPEVATRLAATDYVDVQISLDGATAEVNDAIRG
Rv0693|M.tuberculosis_H37Rv ATAHHVGVKFSTNGVRITPEVATRLAATDYVDVQISLDGATAEVNDAIRG
MAV_4478|M.avium_104 ATAHHVGVKFSTNGVRITPEVAARLAASDYVDVQISLDGATAEVNDAVRG
MMAR_1021|M.marinum_M ATAHHVGVKFSTNGVRINPEVAARLAASDYVDVQISLDGATAEVNDAVRG
MUL_0773|M.ulcerans_Agy99 ATAHHVGVKFSTNGVRINPEVAARLAASDCVDVQISLDGATAEVNDAVRG
Mflv_5060|M.gilvum_PYR-GCK ATEHHVGVKFSTNGVRIDKTVAAKLAASDYVDVQISLDGATAEVNDAVRG
Mvan_1296|M.vanbaalenii_PYR-1 ATEHHVGVKFSTNGVRITPEVAAKLAASDYVDVQISLDGATAEVNDTVRG
TH_1901|M.thermoresistible__bu ATAHNVGVKFSTNGVRITPEVARRLAASDYVDVQISLDGATAEINDAVRG
MAB_3835c|M.abscessus_ATCC_199 ATEHHVGVKFSTNGVRITEEVAARLAASDYVDVQISLDGATAEVNDAVRG
MSMEG_3721|M.smegmatis_MC2_155 ANGLGMYTNLITSGLGFSTRRAEQLRDAGLDHVQVSIQADQPEVSDRIAG
*. : .:: *.*: : * :* :. .**:*::. .*:.* : *
Mb0712|M.bovis_AF2122/97 TGSFDMAVRALQNLAAAGFA---GVKISVVITRRNVAQLDEFATLASRYG
Rv0693|M.tuberculosis_H37Rv TGSFDMAVRALQNLAAAGFA---GVKISVVITRRNVAQLDEFATLASRYG
MAV_4478|M.avium_104 PGSFDMAVRALQNLADAGFTQNGGAKISVVVTRHNVGQLDEFAALASRYG
MMAR_1021|M.marinum_M AGSFAMAVRALENLAAAGFA---DAKISVVVTRHNVGQLDDFAALADRYG
MUL_0773|M.ulcerans_Agy99 AGSFAMAVRALENLAAAGFA---DAKISVVVTRHNVGQLDDFAALADRYG
Mflv_5060|M.gilvum_PYR-GCK PGSFAMAVRALENLKEAGFK---DAKISVVVTRHNIDQLDDFKALADTYG
Mvan_1296|M.vanbaalenii_PYR-1 PGSFAMAVRALENLKAAGFK---DAKISVVVTRHNIDQLDDFKALADTYG
TH_1901|M.thermoresistible__bu AGSFDMAVRALQNLADAGFA---DAKISVVVTRHNVDQLDDFKALADRYG
MAB_3835c|M.abscessus_ATCC_199 AGSFDMAVRALENLAAAGFK---DAKISVVVTRHNVDQLDEFASLAARYG
MSMEG_3721|M.smegmatis_MC2_155 TASFERKHDVARLVKKLGWP----LTLNVVLHRQNIDRIGAILDMAAVME
..** . . : *: .:.**: *:*: ::. : :*
Mb0712|M.bovis_AF2122/97 AT-LRITRLRPSGRGTDVWADLHPTADQQVQLYDWLVSKGERVLTGDSFF
Rv0693|M.tuberculosis_H37Rv AT-LRITRLRPSGRGTDVWADLHPTADQQVQLYDWLVSKGERVLTGDSFF
MAV_4478|M.avium_104 AT-LRITRLRPSGRGADVWDELHPTADQQVQLYDWLVAKGERVLTGDSFF
MMAR_1021|M.marinum_M AT-LRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFF
MUL_0773|M.ulcerans_Agy99 AT-LRITRLRPSGRGADVWEELHPTAAQQVALYDWLVAKGERVLTGDSFF
Mflv_5060|M.gilvum_PYR-GCK AT-LRITRLRPSGRGADVWDDLHPTADQQVQLYNWLVAHGERVLTGDSFF
Mvan_1296|M.vanbaalenii_PYR-1 AT-LRITRLRPSGRGADVWDELHPTPAQQVQLYDWLVAHGERVLTGDSFF
TH_1901|M.thermoresistible__bu AT-LRITRLRPSGRGADVWDELHPTAEQQVQLYNWLVAHGEKVLTGDSFF
MAB_3835c|M.abscessus_ATCC_199 AT-LRITRLRPSGRGADVWDELHPTAAQQRQLYDWLVRNGERVLTGDSFF
MSMEG_3721|M.smegmatis_MC2_155 ADRIELANTQYYGWAGRNRAELLPSRDQ--LERAQTVVRAARGRLGNRME
* :.::. : * . :* *: * * .. : *: :
Mb0712|M.bovis_AF2122/97 HLAPLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDH---FLAG
Rv0693|M.tuberculosis_H37Rv HLAPLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDH---FLAG
MAV_4478|M.avium_104 HLAPLGQ-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDR---FLAG
MMAR_1021|M.marinum_M HLAPLGS-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDR---FLAG
MUL_0773|M.ulcerans_Agy99 HLAPLGS-SGALAGLNMCGAGRVVCLIDPVGDVYACPFAIHDR---FLAG
Mflv_5060|M.gilvum_PYR-GCK HLSGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDK---FLAG
Mvan_1296|M.vanbaalenii_PYR-1 HLSGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDK---FLAG
TH_1901|M.thermoresistible__bu HLSGLGE-PGALAGLNLCGAGRVVCLIDPVGDVYACPFAIHDR---FLAG
MAB_3835c|M.abscessus_ATCC_199 HLSAYGDGSGGLPGLNMCGAGRVVCLIDPVGDVYACPFAIHDR---FLAG
MSMEG_3721|M.smegmatis_MC2_155 IIYVIPDYYEKYPKPCMAGWARRQFTVVPNGEALPCPAAHELARGLSLPG
: . . :.* .* : * *:. .** * . *.*
Mb0712|M.bovis_AF2122/97 NVLSDGGFQNVWKNSSLFRELR-EPQSAGACGSCGHYDSCRGGCMAAKFF
Rv0693|M.tuberculosis_H37Rv NVLSDGGFQNVWKNSSLFRELR-EPQSAGACGSCGHYDSCRGGCMAAKFF
MAV_4478|M.avium_104 NVLSDGGFDNVWKNAPLFRELR-EPQSAGACGSCGHYDSCRGGCMAAKFF
MMAR_1021|M.marinum_M NVLTDGGFDQVWKNAPLFRQLR-EPQSAGACGSCGHYDSCRGGCMAAKFF
MUL_0773|M.ulcerans_Agy99 NVLTDGGFDQVWKNAPLFRQLR-EPQSAGACGSCGHYDSCRGGCMAAKFF
Mflv_5060|M.gilvum_PYR-GCK NILTSNGFEDVWQNSELFRELR-EPQSAGACSGCNHYDACRGGCMAAKFF
Mvan_1296|M.vanbaalenii_PYR-1 NVLSDGGFDTVWKHSELFRELR-EPQSAGACSGCGHYDACRGGCMAAKFF
TH_1901|M.thermoresistible__bu NVLTDGGFDNVWKNSQLFRELR-EPQSAGACSGCSHFDSCRGGCMAAKFF
MAB_3835c|M.abscessus_ATCC_199 NVLSDNGFDQVWKHSQLFSELR-EPQSAGACGSCGHYDSCRGGCMAAKFF
MSMEG_3721|M.smegmatis_MC2_155 TNVRRHSLEWIWMQSPLFRQFRGDDWMPDPCRTCDRREIDFGGCRCQAFA
. : .:: :* :: ** ::* : ...* *.: : *** . *
Mb0712|M.bovis_AF2122/97 TGLPLDGPDPECVQGHSEPALARERHLPRPRADHSRGRRV----SKPVPL
Rv0693|M.tuberculosis_H37Rv TGLPLDGPDPECVQGHSEPALARERHLPRPRADHSRGRRV----SKPVPL
MAV_4478|M.avium_104 TGLPLDGPDPECVQGHSAAALAGDREAPRPRADHSRGKRI----RQPVPL
MMAR_1021|M.marinum_M TGLPLDGPDPECVQGYGAPALAQERHAPRPRVDHSRGSRV----SAPVPL
MUL_0773|M.ulcerans_Agy99 TGLPLDGPDPECVQGYGAPALAQERHAPRPRVDHSRGSRE----------
Mflv_5060|M.gilvum_PYR-GCK TGLPMDGPDPECVQGYGEPALAADRDKPKSSVDHSRSGGRG-KPKGPIPL
Mvan_1296|M.vanbaalenii_PYR-1 TGLPMDGPDPECVQGYGEPALALDRDKPRSSVDHSRSGGR--TPKGPIPL
TH_1901|M.thermoresistible__bu TGLPLDGPDPECVEGHGAPALARERTKPKPRVDHSR--------GGPVAL
MAB_3835c|M.abscessus_ATCC_199 TGLPMDGPDPECVQGYGEAALAADRVVPKPSGDHSHSKSKRGPATGPVAL
MSMEG_3721|M.smegmatis_MC2_155 LTGDAARTDPVCHLSPDHGLVETAVAAANDP-------------TLPHDG
.** * . . : ..
Mb0712|M.bovis_AF2122/97 TLSMRP--------PKRPCNESPV-------
Rv0693|M.tuberculosis_H37Rv TLSMRP--------PKRPCNESPV-------
MAV_4478|M.avium_104 TLSVRPPVLTPAAPPARLCNESPV-------
MMAR_1021|M.marinum_M TLTTRP--------PARLCNESPV-------
MUL_0773|M.ulcerans_Agy99 -------------------------------
Mflv_5060|M.gilvum_PYR-GCK TLLSAP--------PKKFCNESPV-------
Mvan_1296|M.vanbaalenii_PYR-1 TLLSAP--------PKKFCNESPV-------
TH_1901|M.thermoresistible__bu KLLTKP--------PARLCNESPV-------
MAB_3835c|M.abscessus_ATCC_199 TLTRRP--------PSRACDENPLSTLRAGR
MSMEG_3721|M.smegmatis_MC2_155 VMIPRPN-------PGRTRTQLPLEVRR---