For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LPTRSRLRSWSPDSLTVAAASLRASGTEFYETVRELHDGIDRMDEAETWQGSAHTAAAAMFGRATDRASA FKDYTERAAATLQRGGSLISNYRTQVQSKADEIDAGPLNVSDQWVVFIDPAGMSAERAAELEAQANAVQA ELNPLLAALDEADSTASTSLLATQATEGISPDLPVKPPIPLTRPPGDEVPSPDTEEGRQFQEIARSQDMA TTVRDITDIEDRDGNRVTTYTMMDGSTQVATEFIDQGLPSQQVYPAGTTRVLHSDKNGNWISETMTTPLE DGGIKTNVSWADGTTITMTETADGVRTGSCITADGRESVLPDEFFTDPLPTLAAGALSGLESKAGQGIPA LTSAELDKVKTGAKWGGPAIGLATMAYNMVSAETLHDACVAGVSGTFGTVASIATAAVGGAAFGPAGAFV GSLGGSWAFGYVGSVVGEVLCPK*
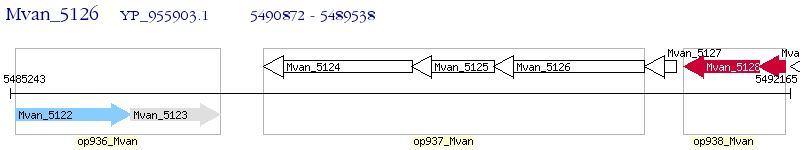
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5126 | - | - | 100% (444) | hypothetical protein Mvan_5126 |
| M. vanbaalenii PYR-1 | Mvan_1063 | - | 5e-08 | 57.78% (45) | hypothetical protein Mvan_1063 |
| M. vanbaalenii PYR-1 | Mvan_5711 | - | 3e-05 | 25.93% (162) | hypothetical protein Mvan_5711 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0376 | - | 4e-08 | 26.63% (169) | hypothetical protein MAB_0376 |
| M. marinum M | MMAR_0482 | - | 3e-10 | 28.14% (199) | hypothetical protein MMAR_0482 |
| M. avium 104 | MAV_0692 | - | 4e-06 | 25.30% (166) | hypothetical protein MAV_0692 |
| M. smegmatis MC2 155 | MSMEG_0365 | - | 4e-08 | 24.88% (201) | hypothetical protein MSMEG_0365 |
| M. thermoresistible (build 8) | TH_3598 | - | 1e-86 | 51.27% (316) | PUTATIVE putative hypothetical protein Mvan_5126 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0482|M.marinum_M MAASGIPDLSALLAWPTDHLTEAAAHWDNVG----EHTYAVAHQLWRDAT
MAV_0692|M.avium_104 MPAT----LSQIQAWSTEHLIDAAAYWTQTA----DRWEDAFLTMRNQAQ
Mvan_5126|M.vanbaalenii_PYR-1 ----MLPTRSRLRSWSPDSLTVAAASLRASGTEFYETVRELHDGIDRMDE
TH_3598|M.thermoresistible__bu -------------------------------------VSRLDDRIDAMPE
MSMEG_0365|M.smegmatis_MC2_155 ----MPGSIFEVRSSDPDALVRAGADIGNRAGRLQAQMDRQQAVIDDLRR
MAB_0376|M.abscessus_ATCC_1997 ----MRPTISQLRAWDVANLDAAAKAIEDASKNLDLSIDGVVRDLD--SA
:
MMAR_0482|M.marinum_M SIDWSGHAAGALRAATHADLQTTSAAVDQLQEAARVARNGASDLYAARSR
MAV_0692|M.avium_104 SITWHGAGGDALRQRTGADVSVVSGKADQLRQAAGIARNGASEISAAQRR
Mvan_5126|M.vanbaalenii_PYR-1 AETWQGSAHTAAAAMFGRATDRASAFKDYTERAAATLQRGGSLISNYRTQ
TH_3598|M.thermoresistible__bu SRTWKGAAHDAATEMFRRATKRSSSFKTYAEAFAGALRSGSSAIGKARTE
MSMEG_0365|M.smegmatis_MC2_155 G--WNGASADAAIESAERSLQHMRRLHDSLTTLQSALQTGGADLAQQRDR
MAB_0376|M.abscessus_ATCC_1997 ESFWKGKAFQSAYDQASNEQSSTNRLSFAISNVAEAVRGGHSALTAARDK
* * . : . : * : : : .
MMAR_0482|M.marinum_M VRYAVTDAQAAGFQVEED-------LSVTDRMTGGSAAQRAARQAQAQAF
MAV_0692|M.avium_104 VLYGVEDAQNAGFTVGED-------LSVTDTRS-TSPAERAARQAQAEAF
Mvan_5126|M.vanbaalenii_PYR-1 VQSKADEIDAGPLNVSDQ-------WVVFIDPAGMSAERAAELEAQANAV
TH_3598|M.thermoresistible__bu LLARADDIDAGPLRVTDQ-------WVVMIDPAEMSEEEAAQLQKQASEM
MSMEG_0365|M.smegmatis_MC2_155 IVNHVDQLHAQGWQVADDGTVSIRPGSVLDFFAKVSPVQRVMLEALAARA
MAB_0376|M.abscessus_ATCC_1997 VMLLVDEATSKGLHVADDG-------------TVTAPEGNRELQAEARRL
: . : * :: : : : *
MMAR_0482|M.marinum_M AGDLRQRAAQLVGLDAQVATKVTAAVAGIGSSFPQIPAPAAPPMGNPVRA
MAV_0692|M.avium_104 AADLRLRAEQLEAADTKAAGQITATAGDVGN-----ATFTPAHNGNGVQL
Mvan_5126|M.vanbaalenii_PYR-1 QAELNPLLAALDEADSTASTSLLATQATEGIS-PDLPVKPPIPLTRPPGD
TH_3598|M.thermoresistible__bu QTEINELLLAVGEADDWTALQLTAAAAGKGAAFANLAYSAPSIVEPAPRD
MSMEG_0365|M.smegmatis_MC2_155 SAELKAMLAQFDATDVTVGAQIRDAVAGLTAHGPAPASPPPNNAKALVDR
MAB_0376|M.abscessus_ATCC_1997 TDAIRQALKSLEEADRTSSNDLRKASASVDAEHNSASTQPAAFMPASSGG
:. . * . .: : . . ..
MMAR_0482|M.marinum_M VDNRTFKQNP------------PPAPVDPKDMTAEEARAAWADINSEIGK
MAV_0692|M.avium_104 VD---FKQDG------------GPVCDDPD--------------------
Mvan_5126|M.vanbaalenii_PYR-1 EVPSPDTEEG------------RQFQEIAR---SQDMATTVRDITDIEDR
TH_3598|M.thermoresistible__bu EVPDPSTEDG------------KLFQEIAR---AQDMATTIREVSETTDR
MSMEG_0365|M.smegmatis_MC2_155 LAGMTPDERREFLAGLSPETVHEMVLADPETMGNTDGVPFETRIEANDIN
MAB_0376|M.abscessus_ATCC_1997 AASGSMKAGS------------GGFSGGGFG-----GGSGGGHGGGFGGG
MMAR_0482|M.marinum_M YNARCGRTFVLPTEQGAYDACIADKGP-------------------LLDR
MAV_0692|M.avium_104 YNDRFFRKFMGRVGAGAAIGGLAG--------------------------
Mvan_5126|M.vanbaalenii_PYR-1 DGNRVTTYTMMDGSTQVATEFIDQG---------------------LPSQ
TH_3598|M.thermoresistible__bu YGNAVTTFHMVDGSEHIRTDWIDQG---------------------LPSQ
MSMEG_0365|M.smegmatis_MC2_155 IRNALEEELAKPSPDQARVQQLQAMLTPIKDPAGSGKLVPRKFVAFSPDG
MAB_0376|M.abscessus_ATCC_1997 HGGGGGGGHSVPDGSYSNTGAPSLR-------------------------
MMAR_0482|M.marinum_M QAAIRARLGQLGIPVEGEDPTHPGTAEPPFPPPTRINGLTEHGAQR----
MAV_0692|M.avium_104 --APEAGVGAIPGAIIGG-------------AAGGIEAFIEGATES----
Mvan_5126|M.vanbaalenii_PYR-1 QVYPAGTTRVLHSDKNGNWISETMTTPLEDGGIKTNVSWADGTTIT----
TH_3598|M.thermoresistible__bu QIHPAGTIQILHVDKNKNWISDTTSMPREDGGNLTTVSWADGTTIA----
MSMEG_0365|M.smegmatis_MC2_155 NGRMIEMIGEIKPGIKGVGVVVPGTNTNLNGSGSNHTSAVELSKQSGSPV
MAB_0376|M.abscessus_ATCC_1997 PSGPSVFMNLDAGQMEVARKIIEEGLRRGLSPEAIQIALATALTESG---
.
MMAR_0482|M.marinum_M --------------------------------------------------
MAV_0692|M.avium_104 --------------------------------------------------
Mvan_5126|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3598|M.thermoresistible__bu --------------------------------------------------
MSMEG_0365|M.smegmatis_MC2_155 FLYMDGDFPQGLDKAADPAYAASMAPKLVSFGHEIDREVGIGAPDTPVTY
MAB_0376|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0482|M.marinum_M -------------------------VEGRDGHGVNDNAMQDAVAHPIEPP
MAV_0692|M.avium_104 -------------------------EPPKCGK------------------
Mvan_5126|M.vanbaalenii_PYR-1 -------------------------MTETADGVRTGSCITADGRESVLPD
TH_3598|M.thermoresistible__bu -------------------------ISQNPDGTRTGSCTTKTDY-AILPD
MSMEG_0365|M.smegmatis_MC2_155 IGHSYGGAIVGTAEQMGLRADRVLHASSAGTGILSGDYTNPNPNVQRFSM
MAB_0376|M.abscessus_ATCC_1997 -------------------------LRSLANSSVPDSMMFPNDGVGHDHD
MMAR_0482|M.marinum_M QFMPDQYGGTWRYIGKDATVSLNENGQVTTAWANSRNGWRNP--------
MAV_0692|M.avium_104 --------------------------------------------------
Mvan_5126|M.vanbaalenii_PYR-1 EFFTDPLP-TLAAGALSGLESKAGQGIPALTSAELDKVKTGAKWGGPAIG
TH_3598|M.thermoresistible__bu SFSTTQFR-TWQVPRSPDWRSRPGKGS-----------------------
MSMEG_0365|M.smegmatis_MC2_155 TAPGDPISLVQSLPRDVAISSIPGVDSIPGIPHNTDGRIGNPLGGLPSAT
MAB_0376|M.abscessus_ATCC_1997 SVGPFQQRQSWGATADLMDPTKSAGKFYDQLVKVSGWQDMSVAQAAQSVQ
MMAR_0482|M.marinum_M --------------------------------------------------
MAV_0692|M.avium_104 --------------------------------------------------
Mvan_5126|M.vanbaalenii_PYR-1 LATMAYNMVSAETLHDACVAGVSGTFGTVASIATAAVGGAAFGPAGAFVG
TH_3598|M.thermoresistible__bu -------MV-----------------------------------------
MSMEG_0365|M.smegmatis_MC2_155 DPDNIPGVTRLDTGYYGAEGAHPNQVIVGPDGHGKYWDDYDSDAFKNMVA
MAB_0376|M.abscessus_ATCC_1997 RSAFPDAYAKYEAQAAQIFHQVTGR-------------------------
MMAR_0482|M.marinum_M --------------------------------------------------
MAV_0692|M.avium_104 --------------------------------------------------
Mvan_5126|M.vanbaalenii_PYR-1 SLGGSWAFGYVGSVVGEVLCPK----------------------------
TH_3598|M.thermoresistible__bu --------------------------------------------------
MSMEG_0365|M.smegmatis_MC2_155 VISGGEATGYVERGIETNYVDIDLGDDGDFRDEAADQAKAGAAPKAPRLP
MAB_0376|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0482|M.marinum_M --------------------------------
MAV_0692|M.avium_104 --------------------------------
Mvan_5126|M.vanbaalenii_PYR-1 --------------------------------
TH_3598|M.thermoresistible__bu --------------------------------
MSMEG_0365|M.smegmatis_MC2_155 WDDNGTYERRQHPWDNPRVTDNPGLGPRVPVR
MAB_0376|M.abscessus_ATCC_1997 --------------------------------