For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AIRASREVLIEAPRDAVMEALADVGALPSYSSMHKRAEVMDRYPDGRPHHVRVKIRLLGLVDTEILEYRW GPDWLVWDAQPTSQQYAQHVEYRLSSDDHGECTRVRIDITVEPVVPIPEFLIRRACGGVLDAATTGLREL ALRDAGT*
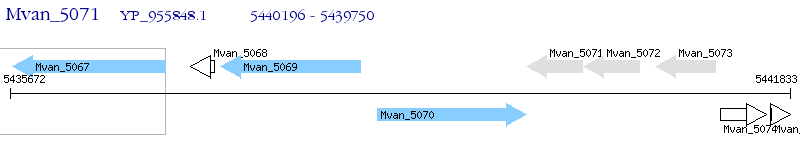
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5071 | - | - | 100% (148) | cyclase/dehydrase |
| M. vanbaalenii PYR-1 | Mvan_5072 | - | 2e-41 | 53.42% (146) | cyclase/dehydrase |
| M. vanbaalenii PYR-1 | Mvan_5073 | - | 2e-27 | 38.19% (144) | cyclase/dehydrase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0880 | - | 4e-46 | 60.00% (140) | hypothetical protein Mb0880 |
| M. gilvum PYR-GCK | Mflv_1679 | - | 9e-41 | 53.15% (143) | cyclase/dehydrase |
| M. tuberculosis H37Rv | Rv0857 | - | 4e-46 | 60.00% (140) | hypothetical protein Rv0857 |
| M. leprae Br4923 | MLBr_00889 | - | 4e-08 | 26.06% (142) | hypothetical protein MLBr_00889 |
| M. abscessus ATCC 19977 | MAB_0831 | - | 2e-27 | 37.06% (143) | hypothetical protein MAB_0831 |
| M. marinum M | MMAR_4679 | - | 5e-46 | 57.53% (146) | hypothetical protein MMAR_4679 |
| M. avium 104 | MAV_0977 | - | 3e-49 | 61.81% (144) | cyclase/dehydrase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5736 | - | 4e-26 | 36.81% (144) | cyclase/dehydrase |
| M. thermoresistible (build 8) | TH_3096 | - | 5e-43 | 52.70% (148) | PUTATIVE cyclase/dehydrase |
| M. ulcerans Agy99 | MUL_0299 | - | 3e-44 | 59.18% (147) | hypothetical protein MUL_0299 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1679|M.gilvum_PYR-GCK -------MAVRASSELVIEAPPEAIMDALADIESVVSWSSLHKDAEIVDR
TH_3096|M.thermoresistible__bu -------MAVRASREVVIDAPVEAILDVLADIDQVPSWSTLHRRAEVLDR
Mb0880|M.bovis_AF2122/97 MIANLVAVAIRASREVVIEAPPEVIVEALADMDAVPSWSSVHKRVEVVDT
Rv0857|M.tuberculosis_H37Rv MIANLVAVAIRASREVVIEAPPEVIVEALADMDAVPSWSSVHKRVEVVDT
MMAR_4679|M.marinum_M MVASLVAVAIQASGEIVIDAPPEVIMEALADMGAVPSWSVVHKRVEVVDN
MAV_0977|M.avium_104 -------MAVKASREFVVNAPPEVVMEALTDVGVLSSWSPLHKHIEVIDR
MUL_0299|M.ulcerans_Agy99 -------MAVKAAREFVIDAPPEVVMEALTDVGVLVSWSPLHKSVEVIDY
Mvan_5071|M.vanbaalenii_PYR-1 -------MAIRASREVLIEAPRDAVMEALADVGALPSYSSMHKRAEVMDR
MAB_0831|M.abscessus_ATCC_1997 -------MATSDTREIVIEATPEQIMDVIADFEVLPKWSSAHQSSTVLTT
MSMEG_5736|M.smegmatis_MC2_155 -------MATSDSREVLIEASPEEILDVIADVESTPSWSPQYTHAEITER
MLBr_00889|M.leprae_Br4923 -------MADKTTQTFYIDANPGEVMKTIADIESYPQWISEYKEVEVLEV
:* : . ::* ::..::*. .: : :
Mflv_1679|M.gilvum_PYR-GCK HPDGRPHHVRATVKIMGLSDQEMLEYHWGDDW--VVWDAEQTSRQRCQHG
TH_3096|M.thermoresistible__bu HPDGRPHHVRATLRILGVTDRQILEYHWGPSW--MVWDAEETFQQRGQHG
Mb0880|M.bovis_AF2122/97 YSDGRPHHVKVTIKVAGIVDTELLEYHWGPDW--VVWDAAKTAQQHGQHG
Rv0857|M.tuberculosis_H37Rv YSDGRPHHVKVTIKVAGIVDTELLEYHWGPDW--VVWDAAKTAQQHGQHG
MMAR_4679|M.marinum_M YPDGRPHHVRVTIKVTGIADTELLEYHWGPDW--MVWDAQRTVQQHGQHG
MAV_0977|M.avium_104 YPDGRPHHVKTTIKILGLVDKEILEYHWGPDW--VVYDAKGTSQQHGQHV
MUL_0299|M.ulcerans_Agy99 YPDGRPHHVKATIKILGLVDKEILEYHWGPDW--VCWDADQTFQQRGQHV
Mvan_5071|M.vanbaalenii_PYR-1 YPDGRPHHVRVKIRLLGLVDTEILEYRWGPDW--LVWDAQPTSQQYAQHV
MAB_0831|M.abscessus_ATCC_1997 GEDGRPHQVTMKVKTAGITDEQTVAYTWGPDT--VSWTLVKASQQRRQDG
MSMEG_5736|M.smegmatis_MC2_155 YDDGRPRRAKMTVKAAGLTDDQVIEYTWSENK--VSWTLVSAGQLKAQDA
MLBr_00889|M.leprae_Br4923 DDEDFPKRARMLMDAKIFKDTLIMSYDWTADHQSVSWILESSSLLKSLEG
:. *::. : . * : * * . : : : .
Mflv_1679|M.gilvum_PYR-GCK EYNLTPVGEER-TRVRFDLILDLAAPYPTFLVRRAKQMVLDIALQNLRQR
TH_3096|M.thermoresistible__bu EYNLTPVGQDR-TRVRFDLIMELAAPVPHFLIKRGRRIVLDVATEKLKER
Mb0880|M.bovis_AF2122/97 EYNLRREDNDK-TRVRFTLTVEPSAPLPAFWVNIARKKILHAATEGLRKQ
Rv0857|M.tuberculosis_H37Rv EYNLRREDNDK-TRVRFTLTVEPSAPLPAFWVNIARKKILHAATEGLRKQ
MMAR_4679|M.marinum_M EYNLRREADNK-TRVRFSITVEPSAPLPEFWVNRARRKILHSALEGLRKR
MAV_0977|M.avium_104 EYNLKPEGVGQ-TRVRFDVTVEPGRPMPAFIVRRASENVLDAAMKGLQEL
MUL_0299|M.ulcerans_Agy99 EYTVRPEGLDK-SRVRFDITVEPSGPISGFIVKRASEHVLDAAAKGLSEL
Mvan_5071|M.vanbaalenii_PYR-1 EYRLSSDDHGECTRVRIDITVEPVVPIPEFLIRRACGGVLDAATTGLREL
MAB_0831|M.abscessus_ATCC_1997 KYTFTPQ-GDK-THVKFELTVDPLIPLPGFVLKRAIKGAMETATDGLRKQ
MSMEG_5736|M.smegmatis_MC2_155 SYTLTPQ-GDK-TKVRFDISIDLSVPLPGFIVKRTMKGGVETATDGLRKQ
MLBr_00889|M.leprae_Br4923 SYRLVPKGSTT--EVTYELAVDFAIPMIGMLKRKAEHRLIDGALKDLKKR
.* . .* : :: * : . :. * * :
Mflv_1679|M.gilvum_PYR-GCK VL-----AATQ---
TH_3096|M.thermoresistible__bu VYRCLGSSAPKRT-
Mb0880|M.bovis_AF2122/97 VVGRRRFTSG----
Rv0857|M.tuberculosis_H37Rv VVGRRRFTSG----
MMAR_4679|M.marinum_M VMGPAD--------
MAV_0977|M.avium_104 VMRAKDLRDSGEAE
MUL_0299|M.ulcerans_Agy99 VAN-RDATDQAK--
Mvan_5071|M.vanbaalenii_PYR-1 ALR-----DAGT--
MAB_0831|M.abscessus_ATCC_1997 VLATYR--------
MSMEG_5736|M.smegmatis_MC2_155 VLKVKKG-------
MLBr_00889|M.leprae_Br4923 VEG-----------
.