For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATSDSREVLIEASPEEILDVIADVESTPSWSPQYTHAEITERYDDGRPRRAKMTVKAAGLTDDQVIEYTW SENKVSWTLVSAGQLKAQDASYTLTPQGDKTKVRFDISIDLSVPLPGFIVKRTMKGGVETATDGLRKQVL KVKKG*
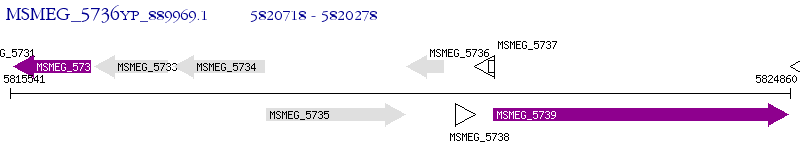
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5736 | - | - | 100% (146) | cyclase/dehydrase |
| M. smegmatis MC2 155 | MSMEG_4251 | - | 1e-25 | 35.46% (141) | cyclase/dehydrase |
| M. smegmatis MC2 155 | MSMEG_0129 | - | 3e-13 | 27.74% (137) | cyclase/dehydrase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0877 | - | 3e-48 | 58.22% (146) | hypothetical protein Mb0877 |
| M. gilvum PYR-GCK | Mflv_1678 | - | 2e-65 | 78.08% (146) | cyclase/dehydrase |
| M. tuberculosis H37Rv | Rv0854 | - | 3e-48 | 58.22% (146) | hypothetical protein Rv0854 |
| M. leprae Br4923 | MLBr_00889 | - | 7e-22 | 31.69% (142) | hypothetical protein MLBr_00889 |
| M. abscessus ATCC 19977 | MAB_0831 | - | 4e-46 | 56.74% (141) | hypothetical protein MAB_0831 |
| M. marinum M | MMAR_4682 | - | 1e-48 | 58.22% (146) | hypothetical protein MMAR_4682 |
| M. avium 104 | MAV_0975 | - | 3e-48 | 58.90% (146) | cyclase/dehydrase superfamily protein |
| M. thermoresistible (build 8) | TH_2031 | - | 4e-53 | 64.29% (140) | cyclase/dehydrase |
| M. ulcerans Agy99 | MUL_0301 | - | 9e-49 | 58.22% (146) | hypothetical protein MUL_0301 |
| M. vanbaalenii PYR-1 | Mvan_5073 | - | 1e-66 | 80.82% (146) | cyclase/dehydrase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1678|M.gilvum_PYR-GCK ----------MATKDSREVVIEATPEQILDVIADVEATPTWSPQYQKAEV
Mvan_5073|M.vanbaalenii_PYR-1 MATSVVYGRAMATKDSREVVIEATPEQILDVIADVEATPTWSPQYQKAEI
MSMEG_5736|M.smegmatis_MC2_155 ----------MATSDSREVLIEASPEEILDVIADVESTPSWSPQYTHAEI
TH_2031|M.thermoresistible__bu ----------MAVSDSRDVVIDATPAQILDVIADVAAVPEWSPQYLSAEV
Mb0877|M.bovis_AF2122/97 ----------MAIKESRDIVIEASPEEILDVIADFEAMTEWSPAHQSVEI
Rv0854|M.tuberculosis_H37Rv ----------MAIKESRDIVIEASPEEILDVIADFEAMTEWSPAHQSVEI
MMAR_4682|M.marinum_M ----------MAIKETRDIVIEASPQEILDVIADFEAMPGWSSPHQSAEI
MUL_0301|M.ulcerans_Agy99 ----------MAIKETRDIVIEASPQEILDVIADFEAMPEWSSPHQSAEI
MAV_0975|M.avium_104 ----------MALKESRDIVIEASPEEILDVIADFESMPQWSEPHQSAEV
MAB_0831|M.abscessus_ATCC_1997 ----------MATSDTREIVIEATPEQIMDVIADFEVLPKWSSAHQSSTV
MLBr_00889|M.leprae_Br4923 ----------MADKTTQTFYIDANPGEVMKTIADIESYPQWISEYKEVEV
** . :: . *:*.* :::..***. . * : :
Mflv_1678|M.gilvum_PYR-GCK LERFDNGRPKQVKMTVKAAGLTDEQVIEYTWGDD--RVTWTLVSASQLKA
Mvan_5073|M.vanbaalenii_PYR-1 LDRYDNGRPKQVRMTVKAAGLTDEQVIEYTWGDN--EVSWTLVSATQLKA
MSMEG_5736|M.smegmatis_MC2_155 TERYDDGRPRRAKMTVKAAGLTDDQVIEYTWSEN--KVSWTLVSAGQLKA
TH_2031|M.thermoresistible__bu LDAHPDGRPRRVKMKVKAAGLTDEQIVDYTWTDH--SAAWTLVSAGQLRS
Mb0877|M.bovis_AF2122/97 LETGDDGRPSKVKMKVKTAGITDEQVVAYSWTDR--SVRWTLVSSTQQRS
Rv0854|M.tuberculosis_H37Rv LETGDDGRPSKVKMKVKTAGITDEQVVAYSWTDR--SVRWTLVSSTQQRS
MMAR_4682|M.marinum_M LETGADGRPSQVKMKVKTAGITDEQVVAYSWGDN--KVNWSLVSSGQQRS
MUL_0301|M.ulcerans_Agy99 FETGADGRPSQVKMKVKTAGITDEQVVAYSWGDN--KVNWSLVSSGQQRS
MAV_0975|M.avium_104 LATGDDGRPSQVRMKVKVAGITDEQVVAYTWADN--VVSWTLVSSSQQKA
MAB_0831|M.abscessus_ATCC_1997 LTTGEDGRPHQVTMKVKTAGITDEQTVAYTWGPD--TVSWTLVKASQQRR
MLBr_00889|M.leprae_Br4923 LEVDDEDFPKRARMLMDAKIFKDTLIMSYDWTADHQSVSWILESSSLLKS
:. * :. * :.. :.* : * * . * * .: :
Mflv_1678|M.gilvum_PYR-GCK QDAGYTLTPEGDKTRVRFDIAIDLSVPLPGFLLKRTMKGGVETATEGLKK
Mvan_5073|M.vanbaalenii_PYR-1 QDAGYTLTPAGDKTKVKFDIAIDLSVPLPGFIIKRTIKGGVETATEGLRK
MSMEG_5736|M.smegmatis_MC2_155 QDASYTLTPQGDKTKVRFDISIDLSVPLPGFIVKRTMKGGVETATDGLRK
TH_2031|M.thermoresistible__bu QDAKYTLRPDGTRTKVTFEITVDPSVPLPGFVLKRAMRGAMETATDGLRK
Mb0877|M.bovis_AF2122/97 QDGKYELTPKGDNTLVQFEITVDPQVPLPGFVLKRAIKGTIDTATEALRS
Rv0854|M.tuberculosis_H37Rv QDGKYELTPKGDNTLVQFEITVDPQVPLPGFVLKRAIKGTIDTATEALRS
MMAR_4682|M.marinum_M QEGSYLLTPEGDKTRVKLELAVDAVVPLPGFVLKRAVKGTVESGLDELRK
MUL_0301|M.ulcerans_Agy99 QEGSYLLTPEGDKTRVKLELAVDAVVPLPGFVLKRAVKGTVESGLDELRK
MAV_0975|M.avium_104 QDGKYTLVPQGDATLVKFELVADPNVPLPGFVLKRAVKGTIDSATKALRQ
MAB_0831|M.abscessus_ATCC_1997 QDGKYTFTPQGDKTHVKFELTVDPLIPLPGFVLKRAIKGAMETATDGLRK
MLBr_00889|M.leprae_Br4923 LEGSYRLVPKGSTTEVTYELAVDFAIPMIGMLKRKAEHRLIDGALKDLKK
:. * : * * * * :: * :*: *:: ::: : :: . . *:.
Mflv_1678|M.gilvum_PYR-GCK QVHKVLKG-
Mvan_5073|M.vanbaalenii_PYR-1 QVLKVIKG-
MSMEG_5736|M.smegmatis_MC2_155 QVLKVKKG-
TH_2031|M.thermoresistible__bu RVEQLYGR-
Mb0877|M.bovis_AF2122/97 QVLKVKKGQ
Rv0854|M.tuberculosis_H37Rv QVLKVKKGQ
MMAR_4682|M.marinum_M QVLKVKKG-
MUL_0301|M.ulcerans_Agy99 QVLKVKKG-
MAV_0975|M.avium_104 RVLEVKKGK
MAB_0831|M.abscessus_ATCC_1997 QVLATYR--
MLBr_00889|M.leprae_Br4923 RVEG-----
:*