For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAVRASSELVIEAPPEAIMDALADIESVVSWSSLHKDAEIVDRHPDGRPHHVRATVKIMGLSDQEMLEYH WGDDWVVWDAEQTSRQRCQHGEYNLTPVGEERTRVRFDLILDLAAPYPTFLVRRAKQMVLDIALQNLRQR VLAATQ
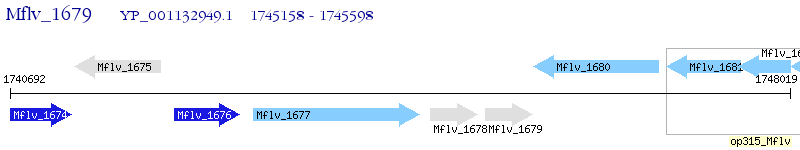
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1679 | - | - | 100% (146) | cyclase/dehydrase |
| M. gilvum PYR-GCK | Mflv_1678 | - | 3e-32 | 43.26% (141) | cyclase/dehydrase |
| M. gilvum PYR-GCK | Mflv_0509 | - | 1e-30 | 45.07% (142) | cyclase/dehydrase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0880 | - | 2e-45 | 54.93% (142) | hypothetical protein Mb0880 |
| M. tuberculosis H37Rv | Rv0857 | - | 2e-45 | 54.93% (142) | hypothetical protein Rv0857 |
| M. leprae Br4923 | MLBr_00889 | - | 2e-15 | 30.77% (143) | hypothetical protein MLBr_00889 |
| M. abscessus ATCC 19977 | MAB_0831 | - | 2e-29 | 41.96% (143) | hypothetical protein MAB_0831 |
| M. marinum M | MMAR_4679 | - | 3e-47 | 55.63% (142) | hypothetical protein MMAR_4679 |
| M. avium 104 | MAV_0978 | - | 1e-50 | 57.75% (142) | hypothetical protein MAV_0978 |
| M. smegmatis MC2 155 | MSMEG_5736 | - | 3e-30 | 42.25% (142) | cyclase/dehydrase |
| M. thermoresistible (build 8) | TH_3096 | - | 2e-57 | 66.67% (141) | PUTATIVE cyclase/dehydrase |
| M. ulcerans Agy99 | MUL_0299 | - | 2e-45 | 54.61% (141) | hypothetical protein MUL_0299 |
| M. vanbaalenii PYR-1 | Mvan_5072 | - | 5e-73 | 86.30% (146) | cyclase/dehydrase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1679|M.gilvum_PYR-GCK -------MAVRASSELVIEAPPEAIMDALADIESVVSWSSLHKDAEIVDR
Mvan_5072|M.vanbaalenii_PYR-1 -------MAVRASSEVVIEAPPEAILDALADIEAVASWSSLHKDAEVVDR
TH_3096|M.thermoresistible__bu -------MAVRASREVVIDAPVEAILDVLADIDQVPSWSTLHRRAEVLDR
Mb0880|M.bovis_AF2122/97 MIANLVAVAIRASREVVIEAPPEVIVEALADMDAVPSWSSVHKRVEVVDT
Rv0857|M.tuberculosis_H37Rv MIANLVAVAIRASREVVIEAPPEVIVEALADMDAVPSWSSVHKRVEVVDT
MMAR_4679|M.marinum_M MVASLVAVAIQASGEIVIDAPPEVIMEALADMGAVPSWSVVHKRVEVVDN
MAV_0978|M.avium_104 -------MAVQASSEIVIDAPPEVIMEALADMDAVPSWSSVHKRVEVIDK
MUL_0299|M.ulcerans_Agy99 -------MAVKAAREFVIDAPPEVVMEALTDVGVLVSWSPLHKSVEVIDY
MAB_0831|M.abscessus_ATCC_1997 -------MATSDTREIVIEATPEQIMDVIADFEVLPKWSSAHQSSTVLTT
MSMEG_5736|M.smegmatis_MC2_155 -------MATSDSREVLIEASPEEILDVIADVESTPSWSPQYTHAEITER
MLBr_00889|M.leprae_Br4923 -------MADKTTQTFYIDANPGEVMKTIADIESYPQWISEYKEVEVLEV
:* : . *:* ::..::*. .* : :
Mflv_1679|M.gilvum_PYR-GCK HPDGRPHHVRATVKIMGLSDQEMLEYHWGDDW--VVWDAEQTSRQRCQHG
Mvan_5072|M.vanbaalenii_PYR-1 YPDGRPHHVKATVKIMGLTDKEFLEYHWGDDW--VVWDAERTSRQRCQHG
TH_3096|M.thermoresistible__bu HPDGRPHHVRATLRILGVTDRQILEYHWGPSW--MVWDAEETFQQRGQHG
Mb0880|M.bovis_AF2122/97 YSDGRPHHVKVTIKVAGIVDTELLEYHWGPDW--VVWDAAKTAQQHGQHG
Rv0857|M.tuberculosis_H37Rv YSDGRPHHVKVTIKVAGIVDTELLEYHWGPDW--VVWDAAKTAQQHGQHG
MMAR_4679|M.marinum_M YPDGRPHHVRVTIKVTGIADTELLEYHWGPDW--MVWDAQRTVQQHGQHG
MAV_0978|M.avium_104 HPDGRPHHVRVTIAVVGIHDTELLEYHWGPDW--MVWDADRTAQQHGQHG
MUL_0299|M.ulcerans_Agy99 YPDGRPHHVKATIKILGLVDKEILEYHWGPDW--VCWDADQTFQQRGQHV
MAB_0831|M.abscessus_ATCC_1997 GEDGRPHQVTMKVKTAGITDEQTVAYTWGPDT--VSWTLVKASQQRRQDG
MSMEG_5736|M.smegmatis_MC2_155 YDDGRPRRAKMTVKAAGLTDDQVIEYTWSENK--VSWTLVSAGQLKAQDA
MLBr_00889|M.leprae_Br4923 DDEDFPKRARMLMDAKIFKDTLIMSYDWTADHQSVSWILESSSLLKSLEG
:. *::. : . * : * * . : * : : .
Mflv_1679|M.gilvum_PYR-GCK EYNLTPVGEERTRVRFDLILDLAAPYPTFLVRRAKQMVLDIALQNLRQRV
Mvan_5072|M.vanbaalenii_PYR-1 EYNLTSVGEQRTRVRFDLLLDLAAPYPGFLVKRAKKMVLDIALQNLRQRV
TH_3096|M.thermoresistible__bu EYNLTPVGQDRTRVRFDLIMELAAPVPHFLIKRGRRIVLDVATEKLKERV
Mb0880|M.bovis_AF2122/97 EYNLRREDNDKTRVRFTLTVEPSAPLPAFWVNIARKKILHAATEGLRKQV
Rv0857|M.tuberculosis_H37Rv EYNLRREDNDKTRVRFTLTVEPSAPLPAFWVNIARKKILHAATEGLRKQV
MMAR_4679|M.marinum_M EYNLRREADNKTRVRFSITVEPSAPLPEFWVNRARRKILHSALEGLRKRV
MAV_0978|M.avium_104 EYNLSRLGDDKTRVRFTITVEPWVPLPEFWVRRARKKILHAALEGLRKRV
MUL_0299|M.ulcerans_Agy99 EYTVRPEGLDKSRVRFDITVEPSGPISGFIVKRASEHVLDAAAKGLSELV
MAB_0831|M.abscessus_ATCC_1997 KYTFTPQG-DKTHVKFELTVDPLIPLPGFVLKRAIKGAMETATDGLRKQV
MSMEG_5736|M.smegmatis_MC2_155 SYTLTPQG-DKTKVRFDISIDLSVPLPGFIVKRTMKGGVETATDGLRKQV
MLBr_00889|M.leprae_Br4923 SYRLVPKG-STTEVTYELAVDFAIPMIGMLKRKAEHRLIDGALKDLKKRV
.* . . :.* : : :: * : . . :. * . * : *
Mflv_1679|M.gilvum_PYR-GCK LAATQ-------
Mvan_5072|M.vanbaalenii_PYR-1 LAGAE-------
TH_3096|M.thermoresistible__bu YRCLGSSAPKRT
Mb0880|M.bovis_AF2122/97 VGRRRFTSG---
Rv0857|M.tuberculosis_H37Rv VGRRRFTSG---
MMAR_4679|M.marinum_M MGPAD-------
MAV_0978|M.avium_104 MVPEAFGRAD--
MUL_0299|M.ulcerans_Agy99 ANRDATDQAK--
MAB_0831|M.abscessus_ATCC_1997 LATYR-------
MSMEG_5736|M.smegmatis_MC2_155 LKVKKG------
MLBr_00889|M.leprae_Br4923 EG----------