For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGSRRDHRDPDGQQGGRYYPPRPPAGEHPGMANYPSDPAMSPGNRRSGRPGPSSQSANRWLPPLDDSSR PHGHSYPPPPGRGADEKVTVTRAAAQRSREMGSKMYGLVHRAATADGADKSGLTALTWPVVANFAVDAAM AVALANTLFFAAASGESKSRVALYLLITIAPFAVIAPLIGPALDRLQHGRRVALAASFSLRTVLAVVLIA NFDSATGSFPSWVLYPCALGMMVLSKSFSVLRSAVTPRVLPPTIDLVRVNSRLTTFGLLGGTMIGGGIAA AAEWGFQLFQMPGALYVVVAVTIGGAVLAMRIPKWVEVTAGEVPTTLSYHGQTEGLRREPHGAVSAKTRQ PLGRNIITALWGNCTVKVMVGFLFLYPAFVAKAHDASGWEQLRILGMIGAAAAVGNFAGNFTAARLKLGH PARLVVRCATAVTVMALATALSGNLLVAAAATLVTSGASAIAKASLDASLQDDLPEESRASAFGRSESLL QLAWVAGGATGVLIYTELYAGFTTITAILILGLAQTVLSYRGESLVPGFGGNRPVLAEQEGVWTDAAVTR E
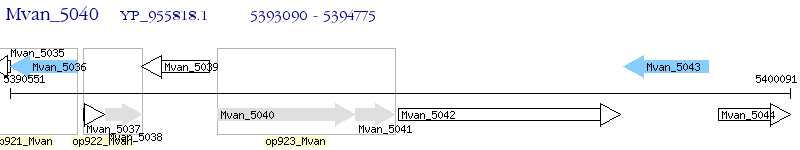
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5040 | - | - | 100% (561) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_1167 | - | 2e-06 | 29.92% (127) | protein of unknown function DUF894, DitE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0900c | - | 0.0 | 74.91% (534) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1711 | - | 0.0 | 88.01% (534) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0876c | - | 0.0 | 75.09% (534) | transmembrane protein |
| M. leprae Br4923 | MLBr_02143 | - | 0.0 | 69.69% (551) | hypothetical protein MLBr_02143 |
| M. abscessus ATCC 19977 | MAB_0875c | - | 0.0 | 70.55% (550) | hypothetical protein MAB_0875c |
| M. marinum M | MMAR_4656 | - | 0.0 | 72.32% (560) | transmembrane protein |
| M. avium 104 | MAV_1005 | - | 0.0 | 71.96% (560) | hypothetical protein MAV_1005 |
| M. smegmatis MC2 155 | MSMEG_5693 | - | 0.0 | 81.42% (506) | transporter, major facilitator family protein |
| M. thermoresistible (build 8) | TH_1066 | - | 0.0 | 79.21% (529) | POSSIBLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_0274 | - | 0.0 | 72.32% (560) | transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5040|M.vanbaalenii_PYR-1 ---MTGSRRDHRDPDGQQGGRYYPPRPPAGEHPGMANYPSDPAMSPGNRR
Mflv_1711|M.gilvum_PYR-GCK ----------------------------------MANYPSDPQTRPGNRR
MSMEG_5693|M.smegmatis_MC2_155 --------------------------------------------------
TH_1066|M.thermoresistible__bu ----------------------------------MANYPSD---DPAPRR
Mb0900c|M.bovis_AF2122/97 --------------MAPTPGRR-TRNGSVNGHPGMANYPPG---DANYRR
Rv0876c|M.tuberculosis_H37Rv --------------MAPTPGRR-TRNGSVNGHPGMANYPPD---DANYRR
MMAR_4656|M.marinum_M ---MSGWRRDHQGRMAASPGRR-TRNGSVSEHPGMANYPTD---GPGNRR
MUL_0274|M.ulcerans_Agy99 ---MSGWRRDHQGRMAASPGRR-TRNGSVSEHPGMANYPTD---GPGNRR
MLBr_02143|M.leprae_Br4923 ---MSGRRRNHPGRLASIPGLR-TRTGSRNQHPGIANYPAD---SSDFRP
MAV_1005|M.avium_104 ---MSGRRQGDPGRVAAKPGRR-PGNSAAAPHPGAANYPAG---DTGDRR
MAB_0875c|M.abscessus_ATCC_199 MRDDYARFGNRHGSSDSPSRRRNQDPRDANSHPGMANYPTG----SAPRE
Mvan_5040|M.vanbaalenii_PYR-1 S------------------GRPGPSSQSANRWLPPLDDSSRPHGH---SY
Mflv_1711|M.gilvum_PYR-GCK P------------------GRPYSPGQAGNRWLPPLDDDPRRAGR---DY
MSMEG_5693|M.smegmatis_MC2_155 -------------------------MPSSNRWLPPLDEESPRAER---TG
TH_1066|M.thermoresistible__bu P------------------GRPKAP-PSANRWLPPLDEDRPTFDRGAGEP
Mb0900c|M.bovis_AF2122/97 S-------------------RRPPPMPSANRYLPPLGEQ-PEPE-----R
Rv0876c|M.tuberculosis_H37Rv S-------------------RRPPPMPSANRYLPPLGEQ-PEPE-----R
MMAR_4656|M.marinum_M T-------------------RRPPPMPSANRYLPPLDRE-PESEDDA-EG
MUL_0274|M.ulcerans_Agy99 T-------------------RRPPPMPSANRYLPPLDRE-PESEDDA-EG
MLBr_02143|M.leprae_Br4923 AQQRRALEQREQQAGQLDPARRSPSMPSANRYLPPLGQQQSEPQ-----H
MAV_1005|M.avium_104 T-------------------RRPPPMPSANRYLPPLGHQ-PQPD----RG
MAB_0875c|M.abscessus_ATCC_199 R---------------------LRASSSSNRYLPPLGEERRERVR-----
:.**:****. .
Mvan_5040|M.vanbaalenii_PYR-1 PP----PPGRG-ADEKVTVTRAAAQRSREMGSKMYGLVHRAATADGADKS
Mflv_1711|M.gilvum_PYR-GCK DSRYHDAPSAA-GGEKVTVTRAAAQRSREMGSKVYGMVHRAATADGADKS
MSMEG_5693|M.smegmatis_MC2_155 SARGAGPTAEPGAGERVTVTRAAAARSREMGSRMYGLVHRAATADGADKS
TH_1066|M.thermoresistible__bu PPRGYRSTGRPRSDERITVTRAAAQRSREMGSRMYGLVHRAATADGADKS
Mb0900c|M.bovis_AF2122/97 SRVPPRTTR---AGERITVTRAAAMRSREMGSRMYLLVHRAATADGADKS
Rv0876c|M.tuberculosis_H37Rv SRVPPRTTR---AGERITVTRAAAMRSREMGSRMYLLVHRAATADGADKS
MMAR_4656|M.marinum_M NEAPPPSPRGLSSGERITVTRAAAMRSREMGSRMYWMVQRAATADGADKS
MUL_0274|M.ulcerans_Agy99 NEAPPPPPRGLSSGERITVTRAAAMRSREMGSRMYWMVQRAATADGADKS
MLBr_02143|M.leprae_Br4923 NSAPPCGPY---PGERIKVTRAAAQRSREMGYRMYWMVQRAATADGADKS
MAV_1005|M.avium_104 AGAPPRGPV---AGERITVTRAAALRSREMGSRMYWMVQRAATADGADKS
MAB_0875c|M.abscessus_ATCC_199 ----RRPPPEQDHPQKLTVTRVAAMRSREMGSRMYGLFQKAATADGADKS
. :::.***.** ****** ::* :.::**********
Mvan_5040|M.vanbaalenii_PYR-1 GLTALTWPVVANFAVDAAMAVALANTLFFAAASGESKSRVALYLLITIAP
Mflv_1711|M.gilvum_PYR-GCK GLTALTWPVVANFAVDAAMAVALANTLFFAAASGESKGKVALYLLITIAP
MSMEG_5693|M.smegmatis_MC2_155 GLTALTWPVVANFAVDAAMAVALANTLFFAAASGESKSRVALYLLITIAP
TH_1066|M.thermoresistible__bu GLTALTWPVVANFAVDAAMAVALANTLFFAAATGESKARVALYLLITIAP
Mb0900c|M.bovis_AF2122/97 GLTALTWPVMANFAVDSAMAVALANTLFFAAASGESKSRVALYLLITIAP
Rv0876c|M.tuberculosis_H37Rv GLTALTWPVMANFAVDSAMAVALANTLFFAAASGESKSRVALYLLITIAP
MMAR_4656|M.marinum_M GLTALTWPTMANFAVDSSMAVALANTLFFAAASGESKSKVALYLLITIAP
MUL_0274|M.ulcerans_Agy99 GLTALTWPTMANFAVDSSMAVALANTLFFAAASGESKSKVALYLLITIAP
MLBr_02143|M.leprae_Br4923 GLTALTWPVVTNFAVDSAMAVALANTLFFAAASGESKSKVALYLLITIAP
MAV_1005|M.avium_104 GLTALTWPVVANFAVDAAMAVALANTLFFAAATGESKGRVALYLLITIAP
MAB_0875c|M.abscessus_ATCC_199 GLTALTYPVMANFAVDAAMAVALANTLFFAAATAESKSKVALYLLITIAP
******:*.::*****::**************:.***.:***********
Mvan_5040|M.vanbaalenii_PYR-1 FAVIAPLIGPALDRLQHGRRVALAASFSLRTVLAVVLIANFDSATGSFPS
Mflv_1711|M.gilvum_PYR-GCK FAVIAPLIGPALDRLQHGRRVALAASFVLRTVLAVVLIANFDSATGSFPS
MSMEG_5693|M.smegmatis_MC2_155 FAVIAPLIGPALDRLQHGRRVALAASFALRTVLIVVLIANYDGATGNFPS
TH_1066|M.thermoresistible__bu FAVIAPLIGPALDRLQHGRRVALATSFGLRTALVVVLIANYDGATGSFPS
Mb0900c|M.bovis_AF2122/97 FAVIAPLIGPALDRLQHGRRVALALSFGLRTALAVVLIMNYDGATGSFPS
Rv0876c|M.tuberculosis_H37Rv FAVIAPLIGPALDRLQHGRRVALALSFGLRTALAVVLIMNYDGATGSFPS
MMAR_4656|M.marinum_M FAVVAPLIGPALDKLQHGRRVALALSFGLRTVLALVLIMNYDGASGSFPS
MUL_0274|M.ulcerans_Agy99 FAVVAPLIGPALDKLQHGRRVALALSFGLRTVLALVLIMNYDGASGSFPS
MLBr_02143|M.leprae_Br4923 FAVIAPLIGPALDRLQHGRRVALATSFVLRTGLATLLIMNYDGATGSYPS
MAV_1005|M.avium_104 FAVIAPLIGPALDRLQHGRRVALAASFVLRTALAAVLIMNYDGASGSYPS
MAB_0875c|M.abscessus_ATCC_199 FAVIAPLIGPALDRLQHGRRAALAASFGLRTLLAVIMIANYDGASGSFPP
***:*********:******.*** ** *** * ::* *:*.*:*.:*.
Mvan_5040|M.vanbaalenii_PYR-1 WVLYPCALGMMVLSKSFSVLRSAVTPRVLPPTIDLVRVNSRLTTFGLLGG
Mflv_1711|M.gilvum_PYR-GCK WVLYPCALGMMVLSKSFSVLRSAVTPRVLPPSIDLVRVNSRLTTFGLLGG
MSMEG_5693|M.smegmatis_MC2_155 WVLYPCALGMMVLSKSFSVLRSAVTPRVLPPTIDLVRVNSRLTTFGLLGG
TH_1066|M.thermoresistible__bu WVLYPCALAMMVLSKSFSVLRSAMTPRVLPPTIDLVRVNSRLTMFGLLGG
Mb0900c|M.bovis_AF2122/97 WVLYPCALAMMVFSKSFSVLRSAVTPRVMPPTIDLVRVNSRLTVFGLLGG
Rv0876c|M.tuberculosis_H37Rv WVLYPCALAMMVFSKSFSVLRSAVTPRVMPPTIDLVRVNSRLTVFGLLGG
MMAR_4656|M.marinum_M WVLYPCALAMMVLSKSFSVLRSAMTPRVMPPSIDLVRVNSRLTVFGLLGG
MUL_0274|M.ulcerans_Agy99 WVLYPCALAMMVLSKSFSVLRSAMTPRVMPPSIDLVRVNSRLTVFGLLGG
MLBr_02143|M.leprae_Br4923 MVLYPCALAMMVLSKSFSVLRSAVTPRVMPPSIDLVRVNSRLTVFGLLGG
MAV_1005|M.avium_104 MVLYPCALAMMVLSKSFSVLRSAVTPRVMPPSIDLVRVNSRLTMFGLLGG
MAB_0875c|M.abscessus_ATCC_199 WVLYPCALSMLVLSKSFGVLRGAVTPRVLPPTIDLVRVNSRLTTFGLVGG
*******.*:*:****.***.*:****:**:*********** ***:**
Mvan_5040|M.vanbaalenii_PYR-1 TMIGGGIAAAAEWGFQ-LFQMPGALYVVVAVTIGGAVLAMRIPKWVEVTA
Mflv_1711|M.gilvum_PYR-GCK TMIGGGIAAAAEYGFQ-LFQMPGALYVVVAVTVAGAVLAMRIPKWVEVTE
MSMEG_5693|M.smegmatis_MC2_155 TVAGGGVAAGAEYLFG-LVHLPGALYVIVAVSVAGAVLSMRIPKWVEVTE
TH_1066|M.thermoresistible__bu TMLGGAVAAAAEYVFN-LFELPGALYVIVAVSLAGAVLSMRIPRWVEVTE
Mb0900c|M.bovis_AF2122/97 TIAGGAIAAGVEFVCTHLFQLPGALFVVVAITIAGASLSMRIPRWVEVTS
Rv0876c|M.tuberculosis_H37Rv TIAGGAIAAGVEFVCTHLFQLPGALFVVVAITIAGASLSMRIPRWVEVTS
MMAR_4656|M.marinum_M TMAGGAVAAGVEFACTRLFELPGALFVVVAVTVAGALLSMRIPRWVEVTA
MUL_0274|M.ulcerans_Agy99 TMAGGAVAAGVEFACTRLFELPGALFVVVAVTVAGALLSMRIPRWVEVTA
MLBr_02143|M.leprae_Br4923 TIAGGAIAAGVEFVCAHLFKLPGALFVVAAITISGALLSMRIPRWVEVTA
MAV_1005|M.avium_104 TIVGGAIAGGVEFVCTHLFKLPGALFVVVAVTVAGASLSMRIPRWVEVTA
MAB_0875c|M.abscessus_ATCC_199 TIVGGAIAAGCELAFTRLLHLPGALFVVIAVTAVGAWASMRIPKWVEVTA
*: **.:*.. * *..:****:*: *:: ** :****:*****
Mvan_5040|M.vanbaalenii_PYR-1 GEVPTTLSYHGQTEGLRREPHGAVSAK-------TRQPLGRNIITALWGN
Mflv_1711|M.gilvum_PYR-GCK GEVPATLSYHGATEMLPREP-AAATGR-------ARQPLGRNIITSLWGN
MSMEG_5693|M.smegmatis_MC2_155 GEVPTTLSYHGGPGELRRPEHPDTTGKPDKP-QSVRQPMGRNTITALWGN
TH_1066|M.thermoresistible__bu GEVPATLTYHG---DLRRHPDQRRHSG------AARQPMGRNTITALWGN
Mb0900c|M.bovis_AF2122/97 GEVPATLSYHRDRGRLRRRWPEEVKNLG----GTLRQPLGRNIITSLWGN
Rv0876c|M.tuberculosis_H37Rv GEVPATLSYHRDRGRLRRRWPEEVKNLG----GTLRQPLGRNIITSLWGN
MMAR_4656|M.marinum_M GEVPATLSYHRDRGRQRRSWPEEVKNLS----GALRQPLGRNIITSLWGN
MUL_0274|M.ulcerans_Agy99 GEVPATLSYHRDRGRQRRSWPEEVKNLS----GALRQPLGRNIITSLWGN
MLBr_02143|M.leprae_Br4923 GEIPATLSYRLHRKPLRQSWPEEVKKVS----GRLRQPLGRNIITSLWGN
MAV_1005|M.avium_104 GEVPATLSYRRDSEPLRRRWPEEVKNVPKKATATLRQPLGRNIITSLWGN
MAB_0875c|M.abscessus_ATCC_199 GEVPTTFGYHGDDGEHPTILLRRSHKQP----EKVRQPLGRNIITSLWGN
**:*:*: *: ***:*** **:****
Mvan_5040|M.vanbaalenii_PYR-1 CTVKVMVGFLFLYPAFVAKAHDASGWEQLRILGMIGAAAAVGNFAGNFTA
Mflv_1711|M.gilvum_PYR-GCK CTVKVMVGFLFLYPAFVAKAHDASGWEQLRILGMIGAAAAIGNFGGNFTA
MSMEG_5693|M.smegmatis_MC2_155 CTIKVMVGFLFLYPAFVAKSHDASGWEQLLILGIIGAAAGIGNFAGNMTA
TH_1066|M.thermoresistible__bu CTIKLMVGFLFLYPAFVAKSHEATGLEQLMILGLIGAAAAVGNFIGNFTA
Mb0900c|M.bovis_AF2122/97 CTIKVMVGFLFLYPAFVAKAHEANGWVQLGMLGLIGAAAAVGNFAGNFTS
Rv0876c|M.tuberculosis_H37Rv CTIKVMVGFLFLYPAFVAKAHEANGWVQLGMLGLIGAAAAVGNFAGNFTS
MMAR_4656|M.marinum_M CTIKVMVGFLFLYPAFVAKAHDANGWVQLGMLGLIGAAAAIGNFAGNFAS
MUL_0274|M.ulcerans_Agy99 CTIKVMVGFLFLYPAFVAKAHDANGWVQLGMLGLIGAAAAIGNFAGNFAS
MLBr_02143|M.leprae_Br4923 CTIKVMVGFLFLYPAFVAKEHQANGWVQLGMLGLIGTAAAIGNFAGNFTS
MAV_1005|M.avium_104 CTIKVMVGFLFLYPAFVAKAHQANGWAQLAMLGMIGAAAGVGNFVGNFTS
MAB_0875c|M.abscessus_ATCC_199 STIKVMVGFLTLYPAFVAKSHDAHGFQQLAMLGMVGAAAGIGNFAGNATS
.*:*:***** ******** *:* * ** :**::*:**.:*** ** ::
Mvan_5040|M.vanbaalenii_PYR-1 ARLKLGHPARLVVRCATAVTVMALATALSGNLLVAAAATLVTSGASAIAK
Mflv_1711|M.gilvum_PYR-GCK ARLKLGHPARLVVRCAIAVTVVALATALTGNLFVAAAATLVTSAASAIAK
MSMEG_5693|M.smegmatis_MC2_155 ARLKLGHPAQMVVRLAIVVTVFALLAALTGKLLVAALATLVTSGASAIAK
TH_1066|M.thermoresistible__bu ARLELGHPAQLVVRAAIAVTIGALATAVTGELLVAALATLITSGASAIAK
Mb0900c|M.bovis_AF2122/97 ARLQLGRPAVLVVRCTVLVTVLAIAAAVAGSLAATAIATLITAGSSAIAK
Rv0876c|M.tuberculosis_H37Rv ARLQLGRPAVLVVRCTVLVTVLAIAAAVAGSLAATAIATLITAGSSAIAK
MMAR_4656|M.marinum_M ARLQLGRPAVLVVRCTVVVTAFALAAAVAGNLVTAAIATLVTSGSSAIAK
MUL_0274|M.ulcerans_Agy99 ARLQLGRPAVLVVRCTMVVTAFALAAAVAGNLVTAAIATLVTSASSAIAK
MLBr_02143|M.leprae_Br4923 ARLQLGRPAVLVVRCTIAVTVLALAASVAGNLLMTTIATLITSGSSAIAK
MAV_1005|M.avium_104 ARLKLGRPAVLVVRCTVAVTAVALAASVAGNLMLAVIATLVTSGASAIAK
MAB_0875c|M.abscessus_ATCC_199 ARLKLGRPALLVVRCASAVTVVALAAAVTGVLAVVVVATLVTSAVSAIAK
***:**:** :*** : ** *: ::::* * .. ***:*:. *****
Mvan_5040|M.vanbaalenii_PYR-1 ASLDASLQDDLPEESRASAFGRSESLLQLAWVAGGATGVLIYTELYAGFT
Mflv_1711|M.gilvum_PYR-GCK ASLDASIQDDLPEESRASGFGRSESLLQLAWVAGGATGVLIYTELYAGFT
MSMEG_5693|M.smegmatis_MC2_155 ASLDASLQDDLPEASRASAFGRSESVLQLAWVAGGAIGVLIYTDLSVGFT
TH_1066|M.thermoresistible__bu ASLDASLQDDLPEQSRASAFGRSESLLQLAWVAGGATGVLVYTELWIGFT
Mb0900c|M.bovis_AF2122/97 ASLDASLQHDLPEESRASGFGRSESTLQLAWVLGGAVGVLVYTELWVGFT
Rv0876c|M.tuberculosis_H37Rv ASLDASLQHDLPEESRASGFGRSESTLQLAWVLGGAVGVLVYTELWVGFT
MMAR_4656|M.marinum_M ASLDASLQHDLPEESRASGFGRSESTLQLAWVLGGAVGVLIYTDLWIGFT
MUL_0274|M.ulcerans_Agy99 ASLDASLQHDLPEESRASGFGRSESTLQLAWVLGGAVGVLIYTDLWIGFT
MLBr_02143|M.leprae_Br4923 ASLDASLQNDLPEESRASGFGRSESTLQLAWVLGGALGVMVYTDLWVGFT
MAV_1005|M.avium_104 ASLDAALQDDLPEESRASGFGRSESTLQLAWVLGGALGVLVYTELWVGFT
MAB_0875c|M.abscessus_ATCC_199 ASLDASLQDDLPEESRASAFGRSEALLQLSWVLGGAIGVMIYTDLWVGFT
*****::*.**** ****.*****: ***:** *** **::**:* ***
Mvan_5040|M.vanbaalenii_PYR-1 TITAILILGLAQTVLSYRGESLVPGFGGNRPVLAEQEGVWTD---AAVTR
Mflv_1711|M.gilvum_PYR-GCK AITAILILGLAQTVLSYRGESLVPGLGGNRPVHAEQEGVRTD---AVVTR
MSMEG_5693|M.smegmatis_MC2_155 TITAVLILGLAQTIVSYRGESLIPGFGGNRPVLAEREGTMAW---GAR--
TH_1066|M.thermoresistible__bu TISAILILGLAQTIVSYRGDSLIPGFGGNRPVLAEPEGRNDH---PAFTP
Mb0900c|M.bovis_AF2122/97 AVSALLILGLAQTIVSFRGDSLIPGLGGNRPVMAEQETTRRG---AAVAP
Rv0876c|M.tuberculosis_H37Rv AVSALLILGLAQTIVSFRGDSLIPGLGGNRPVMAEQETTRRG---AAVAP
MMAR_4656|M.marinum_M TVSALLILGLAQTIVSFRGDSLIPGLGGNRPVMAEQETTRRGSATAGVGP
MUL_0274|M.ulcerans_Agy99 TVSALLILGLAQTIVSFRGDSLIPGLGGNRPVMAEQETTRRGTATAGVGP
MLBr_02143|M.leprae_Br4923 AVSALLILGLAQTVVSFRGDSLIPGLGGNRPVMIEQESMRRA---AAVSP
MAV_1005|M.avium_104 AVTALLILGLAQTLVSFRGNSLIPGLGGNRPIMVEQEGARRGVGRPAVVA
MAB_0875c|M.abscessus_ATCC_199 VISAVLILGLAQTLASFRGVSLIPGLGGNRPVMAEQEGGKVHA-------
.::*:********: *:** **:**:*****: * *
Mvan_5040|M.vanbaalenii_PYR-1 E
Mflv_1711|M.gilvum_PYR-GCK E
MSMEG_5693|M.smegmatis_MC2_155 -
TH_1066|M.thermoresistible__bu R
Mb0900c|M.bovis_AF2122/97 Q
Rv0876c|M.tuberculosis_H37Rv Q
MMAR_4656|M.marinum_M Q
MUL_0274|M.ulcerans_Agy99 Q
MLBr_02143|M.leprae_Br4923 Q
MAV_1005|M.avium_104 E
MAB_0875c|M.abscessus_ATCC_199 -