For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THGFSEFHDELRSVAGDLLAKDRVVDWSVLVDAGWTGLEVPDGLGGSGATFVETAMILDQIGRAASANRY LGSAVLAVGVLNLLEPSEIRDELLTAVASGPGRLGVATDGFAVNGGRLAGRAGFVPDAVGADRLLVLVDG AVAVVDAAQMSVVPQPVVDETRQLATVTAEAAEIAELLTLAGDPASVRHRAEVAIACDSLGLAEQMLAAT VDYVLVRHQFGRPIGSFQAVKHACADMLVAIEVSRRLVSDAVGVLAEGGDAATQAAMAKSHACSAAVDVA GKAMQLHGGIGYTWESGIHVYLKRATLNRALFGSPAAQRKRLSQRYSKGI*
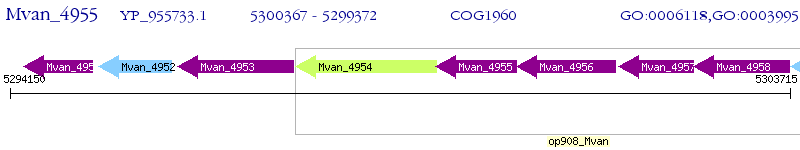
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4955 | - | - | 100% (331) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_0226 | - | 7e-38 | 36.05% (294) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_5289 | - | 5e-37 | 35.91% (298) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1968c | fadE18 | 3e-39 | 38.06% (310) | acyl-CoA dehydrogenase FADE18 |
| M. gilvum PYR-GCK | Mflv_1795 | - | 1e-131 | 72.56% (328) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv1933c | fadE18 | 4e-39 | 38.06% (310) | acyl-CoA dehydrogenase FADE18 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-12 | 31.82% (132) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 3e-33 | 33.12% (314) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_2861 | fadE18 | 3e-39 | 38.11% (307) | acyl-CoA dehydrogenase FadE18 |
| M. avium 104 | MAV_1267 | - | 9e-39 | 35.22% (318) | acyl-CoA dehydrogenase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5618 | - | 3e-92 | 55.88% (340) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2777 | - | 1e-109 | 61.61% (336) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_2893 | fadE18 | 2e-39 | 38.11% (307) | acyl-CoA dehydrogenase FadE18 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1968c|M.bovis_AF2122/97 ----------MDFRYSTEQDDFRASLRGFLGRGAP----VREMAAADGSD
Rv1933c|M.tuberculosis_H37Rv ----------MDFRYSTEQDDFRASLRGFLGRGAP----VREMAAADGSD
MMAR_2861|M.marinum_M ----------MDFRYSTEQDDFRESLRGFLRDQAPPAR-VRQVAAVGGHD
MUL_2893|M.ulcerans_Agy99 ----------MDFRYSTEQDDFRESLRGFLRDQAPPAR-VRQVAAVGGHD
Mvan_4955|M.vanbaalenii_PYR-1 ----------MTHGFSEFHDELRSVAGDLLAKD-----------------
Mflv_1795|M.gilvum_PYR-GCK ----------MRAEFAEFHDELRSVASDLLAKDG----------------
TH_2777|M.thermoresistible__bu ----------MTDQFTEFHDELRAVAAEVLAKD-----------------
MSMEG_5618|M.smegmatis_MC2_155 ------MMTDFTTDFSAIHDELRSVAADLLAKD-----------------
MAV_1267|M.avium_104 ----------MNLELTDEQVALRDTTRRFLAEKAPIAHHVRRLLDDPTGI
MAB_0580|M.abscessus_ATCC_1997 ---MSVPSGLRASGSSEVQEAARDAVRQWARSAAVIES--VRKMESEPDG
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAP---HAADVDQRARF
. : *
Mb1968c|M.bovis_AF2122/97 RRLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPIPFA-
Rv1933c|M.tuberculosis_H37Rv RRLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPIPFA-
MMAR_2861|M.marinum_M KKLWHRLCAELELPALHAPPEYGGAGGTLVETAIAFGELGRALTPVPFA-
MUL_2893|M.ulcerans_Agy99 KKLWHRLCAELELPALHAPPEYGGAGGTLVETAIAFGELGRALTPVPFA-
Mvan_4955|M.vanbaalenii_PYR-1 RVVDWSVLVDAGWTGLEVPDGLGGSGATFVETAMILDQIGRAASANRYLG
Mflv_1795|M.gilvum_PYR-GCK HAPEWRVLADAGWTGLEVPDTLGGAGASFAESAVILDQIGRAAGVTGYLG
TH_2777|M.thermoresistible__bu RTVDWPVLADAGWVGLEVADEFGGAGASFGEVAVICSEMGRAATASSYLG
MSMEG_5618|M.smegmatis_MC2_155 -SVDWPLLVQAGWVGLDAPESLGGAGATFAEVAVICEELGRAAATTGYLG
MAV_1267|M.avium_104 EPAVWRGLADLGCTGLLVPEQHGGAGMTMVEAGIVAEELGAALYPGPWLS
MAB_0580|M.abscessus_ATCC_1997 WGPAYVGLAELGIFGVAVPEAAGGSGAGFDDMIAMVDEAAASLVP-----
MLBr_00737|M.leprae_Br4923 PEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIP
.: .. ** * : .
Mb1968c|M.bovis_AF2122/97 ATVFAIEAILRMGDD-EQRKRLLAGLLTGARIGTIAVSGHDV-AS--ATT
Rv1933c|M.tuberculosis_H37Rv ATVFAIEAILRMGDD-EQRKRLLAGLLTGARIGTIAVSGHDV-AS--ATT
MMAR_2861|M.marinum_M ATILAIEAILRMGDS-EQRKGLLSGLLSGDRIGALAAAGPDL-S---VTS
MUL_2893|M.ulcerans_Agy99 ATILAIEAILRMGDS-EQRKGLLSGLLSGDRIGALAAAGPDL-S---VTS
Mvan_4955|M.vanbaalenii_PYR-1 SAVLAVGVLNLLEPS-EIRDELLTAVASGPGRLGVATD-----------G
Mflv_1795|M.gilvum_PYR-GCK GAVLATGVLNLLRPS-DSRDHLLAAVAGGEPRLAVVTG-----------D
TH_2777|M.thermoresistible__bu GAVLGVGALNAVQPD-AARDDLLTAVAAGAERTALVLG-----------P
MSMEG_5618|M.smegmatis_MC2_155 GAVLGVGALLAVAPN-TSRDNLLTEVVEGRTRVSLALPGDLV-PGDPRVP
MAV_1267|M.avium_104 TAVAAPRALIRLCAGGDTAAGLLRGIADGTTHVAVCFLDTP--------P
MAB_0580|M.abscessus_ATCC_1997 -GPVATSALVAVVLAADGHEELVPPLAAGERTAGLALTG-----------
MLBr_00737|M.leprae_Br4923 AVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMR
: :: :
Mb1968c|M.bovis_AF2122/97 VRAVRRDGRPALTGECTPVLHGHVADLFVVPAVADGS-----IVLHVVAA
Rv1933c|M.tuberculosis_H37Rv VRAVRRDGRPALTGECTPVLHGHVADLFVVPAVADGS-----IVLHVVAA
MMAR_2861|M.marinum_M VRAGRRDGRPVLTGECAPVLHGHVADLFVVPAESDGA-----VILYVVDA
MUL_2893|M.ulcerans_Agy99 VRAGRRDGRPVLTGECAPVLHGHVADLFVVPAESDGA-----VILYVVDA
Mvan_4955|M.vanbaalenii_PYR-1 FAVN----GGRLAGRAGFVPDAVGADRLLVLVD-----------GAVAVV
Mflv_1795|M.gilvum_PYR-GCK FSMS----AGKLTGRAEFVPDAVGADRLLVVTG-----------PEVAVV
TH_2777|M.thermoresistible__bu FTFT----GGRLTGRAEFVPDASGADRLLVLATDS--------DGTPVLT
MSMEG_5618|M.smegmatis_MC2_155 FTVTT---DGRVHGRAAFVPDAGDAHRLLLPARDA--------TGTTVLV
MAV_1267|M.avium_104 MAATREGETTVLHGQARAVPDAAAADVLLALAEDAGD-----TALFGVNT
MAB_0580|M.abscessus_ATCC_1997 ---NLAIADGLASGVLPAVLGGTADGVLLAPVPDG---------WVLVDC
MLBr_00737|M.leprae_Br4923 TRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHK
* : . . .
Mb1968c|M.bovis_AF2122/97 DAPGVTVTPLP----SFDITRPVATLRLAGSPAEPLTAGTPDDMER---V
Rv1933c|M.tuberculosis_H37Rv DAPGVTVTPLP----SFDITRPVATLRLAGSPAEPLTAGTPDDMER---V
MMAR_2861|M.marinum_M AAPGVAVVGLP----SFDITRPVAKLRLVRAPAEALAASSADEVAR---L
MUL_2893|M.ulcerans_Agy99 AAPGVAVVGLP----SFDITRPVAKLRLVRAPAEALAASSADEVAR---L
Mvan_4955|M.vanbaalenii_PYR-1 DAAQMSVVPQP----VVDETRQLATVTAEAAEIAELLTLAGDPAS----V
Mflv_1795|M.gilvum_PYR-GCK ETAAATVTAQP----VVDETRRLATVAADSATVVDILTFDGNPADAVNAL
TH_2777|M.thermoresistible__bu ATAAATVTAQP----VLDETRRLAVVCAEDSGVEAVWRFAGDPRAAARRL
MSMEG_5618|M.smegmatis_MC2_155 D-AAATVIEQP----VLDETRRLAIVEADGAEARDVWPLE-DPHA----L
MAV_1267|M.avium_104 GSAGVAVRPEH----HIDPTHKQSRVTFDAAPARRLATASADDVAA---V
MAB_0580|M.abscessus_ATCC_1997 AEAGVAVEAKK----ATDFSRPWVTVTLHGAAVRPVGLPAAVIADLS---
MLBr_00737|M.leprae_Br4923 DDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALAT
:: . . . :
Mb1968c|M.bovis_AF2122/97 LDVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHACADM
Rv1933c|M.tuberculosis_H37Rv LDVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHACADM
MMAR_2861|M.marinum_M LDVARVLLAAEMLGGAEACLDLAIEYACSRKQFDRPIGSFQAVKHRCADM
MUL_2893|M.ulcerans_Agy99 LDVARVLLAAEMLGGAEACLDLAIEYACSRKQFDRPIGSFQAVKHRCADM
Mvan_4955|M.vanbaalenii_PYR-1 RHRAEVAIACDSLGLAEQMLAATVDYVLVRHQFGRPIGSFQAVKHACADM
Mflv_1795|M.gilvum_PYR-GCK RHRAEIAVACDSLGVAEQMLSDTVDYVKVRHQFGRPIGSFQAVKHACADM
TH_2777|M.thermoresistible__bu ADRAAVAVAIDSLGLAEAMLSATVSYAKVRHQFGRPIGSFQAVKHACADM
MSMEG_5618|M.smegmatis_MC2_155 ARRAALAVACDSLGIAAAMLDATVSYASMRQQFGRPIGSFQAVKHACADM
MAV_1267|M.avium_104 VDDVLIAACADALGAARAVMDLAVEYAKTRTQFGQPIGSFQAVQHLCVDM
MAB_0580|M.abscessus_ATCC_1997 ----VVTLSAEAVGVARWALQTAVEYAKVREQFGKQIGSFQAIKHMCAEM
MLBr_00737|M.leprae_Br4923 LDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADM
: :* * : :: *. * **.. *.:**:::. .:*
Mb1968c|M.bovis_AF2122/97 MIEIDATRATVMFAAMSAAN-------GDELQTVAPLAKAQTAETFVLCA
Rv1933c|M.tuberculosis_H37Rv MIEIDATRATVMFAAMSAAN-------GDELQTVAPLAKAQTAETFVLCA
MMAR_2861|M.marinum_M MIEIDATRAAVMFAQLCAAG-------GDELHATAPLAKAQAADTYVLCA
MUL_2893|M.ulcerans_Agy99 MIEIDATRAAVMFAQLCAAG-------GDELHATAPLAKAQAAGTYVLCA
Mvan_4955|M.vanbaalenii_PYR-1 LVAIEVSRRLVSDAVGVLAE-------GGDAATQAAMAKSHACSAAVDVA
Mflv_1795|M.gilvum_PYR-GCK LVAVEVSRQLVADAVDVLAG-------GGDAAIPAAMAKSYVCAAAVEVA
TH_2777|M.thermoresistible__bu QVQITVARQLVGRAVAALCDPD-----SDDTAAAAAMAKSFVCSAAVEVV
MSMEG_5618|M.smegmatis_MC2_155 LVQVTAARQLIAGAIS------------APDAHTVAKAKSYATEAAVGVA
MAV_1267|M.avium_104 YETVELARSGVIHALWVSDAP------DGDERHLAALRAKAFAGRLAGVA
MAB_0580|M.abscessus_ATCC_1997 LCRVEQADVAVWDAAGCAQDVLANKGDSQQLSLAAAVAAAVAVDAAVANT
MLBr_00737|M.leprae_Br4923 AMKVEAARLIVYAAAARAERG------EPDLGFISAASKCFASDIAMEVT
: : : * . .
Mb1968c|M.bovis_AF2122/97 GSALQIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVK
Rv1933c|M.tuberculosis_H37Rv GSALQIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVK
MMAR_2861|M.marinum_M GSAIQVHGGIAFTWEHDLHLYFRRAKTTEALFGSSAHHRSLLADRVGL--
MUL_2893|M.ulcerans_Agy99 GSAIQVHGGIAFTWEHDLHLYFRRAKTTEALFGSSAHHRSLLADRVGL--
Mvan_4955|M.vanbaalenii_PYR-1 GKAMQLHGGIGYTWESGIHVYLKRATLNRALFGSPAAQRKRLSQRYSKGI
Mflv_1795|M.gilvum_PYR-GCK GKAMQLHGGIGYTWESGIHTYLKRATLNRSLFGSPAAQRRRLSQRYH---
TH_2777|M.thermoresistible__bu GKAMQLHGGIGYTWESGVHVYLKRATLNRALYGSPAAHRSQLVSRYR---
MSMEG_5618|M.smegmatis_MC2_155 GKAMQLHGGIGYTWESGVHTYLKRAALNRSLFGSPREHRTMLSARYRSAP
MAV_1267|M.avium_104 DTAIQVFGGIGYTWEHDAHLYLKRLLGYSAFLGGPDRYLTEVGARLAKRG
MAB_0580|M.abscessus_ATCC_1997 KDCIQILGGIGFTWEHDAHLYLRRAYALQSFVGKPAVWRRKAAALTIGGI
MLBr_00737|M.leprae_Br4923 TDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR--
.:*: *. .:* : . : : *
Mb1968c|M.bovis_AF2122/97 A-------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv A-------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 G-------------------------------------------------
MAV_1267|M.avium_104 SP------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 RRSLTIDLGEVESQRAEVAGQVAEIASAPKAEHQVRLAEAGFLAPHWPAP
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 YGLGAGAAQQLLIDQELHAAGVNRPDLVIGWWAAPTILEHGTPEQIAQFV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 APTLRGEIEWCQLFSEPGAGSDLAALRTKATRVDGGWKLDGQKVWNSKAQ
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 IADWAICLARTNPDVPKHKGITYFLLDMKTPGITVRPLREITGEEMFNEV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 FLDGVIVPDENVVGSVDDGWRLARTTLANERVAMAGGGTLDEAMESLLAL
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 IGDTELDAAQLDTLGAFVCSAQTGALLDLRTALRAIGGQDPGAASSVRKL
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2861|M.marinum_M --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
Mvan_4955|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAV_1267|M.avium_104 --------------------------------------------------
MAB_0580|M.abscessus_ATCC_1997 VGVRHRQGLSEHILDYVDTHGAVESREMWLFMRDRCLSIAGGTTQILLSV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 ----------
Rv1933c|M.tuberculosis_H37Rv ----------
MMAR_2861|M.marinum_M ----------
MUL_2893|M.ulcerans_Agy99 ----------
Mvan_4955|M.vanbaalenii_PYR-1 ----------
Mflv_1795|M.gilvum_PYR-GCK ----------
TH_2777|M.thermoresistible__bu ----------
MSMEG_5618|M.smegmatis_MC2_155 ----------
MAV_1267|M.avium_104 ----------
MAB_0580|M.abscessus_ATCC_1997 AGERLLGLPR
MLBr_00737|M.leprae_Br4923 ----------