For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AADNSSVLVAGARTPIGKFMGSLAGLTGAELGGVAIAAALAKAGISGSQVQYVIMGQVLTAGAGQIPARQ AAVAGGIPMTVPALTVNKVCLSGLDAIALADQLIRAGAFDVVVAGGQESMSQAPHLLENLRAGTKFGDAA LRDHVAVDGLRDVFTDQAMGSLTDSVNALSRADQDEFAAASHRRAAAARKNGVFDGEVVPVVIPQRKGDP VQVTVDEGIRADTTTASLARLRPAFVADGTVTAGNASTMNDGAAAVVVMSKAKATEVGLTWIAEIGAHGV VAGPDSTLQMQPANAIAAACAIEGIDPAALDLVEINEAFAAVGIESTRALGLDPERVNVNGGAIAIGHPL GMSGARIVLHLALELQRRGGGTGAAALCGGGGQGSALIVRC*
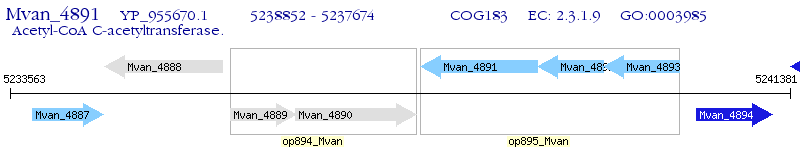
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4891 | - | - | 100% (392) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_4305 | - | e-148 | 69.47% (393) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1976 | - | 8e-61 | 35.86% (396) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1358 | fadA4 | 1e-148 | 69.51% (387) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_2340 | - | 1e-145 | 67.43% (393) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv1323 | fadA4 | 1e-148 | 69.51% (387) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_01158 | fadA4 | 1e-145 | 68.38% (389) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_1463 | - | 1e-159 | 72.59% (394) | acetyl-CoA acetyltransferase FadA |
| M. marinum M | MMAR_4075 | fadA4 | 1e-148 | 69.15% (389) | acetyl-CoA acetyltransferase FadA4 |
| M. avium 104 | MAV_1544 | - | 1e-149 | 69.67% (389) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_4920 | - | 1e-152 | 69.92% (389) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_0255 | - | 8e-57 | 37.73% (387) | acetyl-CoA acetyltransferase |
| M. ulcerans Agy99 | MUL_3938 | fadA4 | 1e-147 | 68.89% (389) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4075|M.marinum_M -------------------MTTSVIVAGARTPIGRLMGSLKDFSASDLGA
MUL_3938|M.ulcerans_Agy99 -------------------MTTSVIVAGARTPIGRLMGSLKDFSASDLGA
Mb1358|M.bovis_AF2122/97 -----------------------MIVAGARTPIGKLMGSLKDFSASELGA
Rv1323|M.tuberculosis_H37Rv -----------------------MIVAGARTPIGKLMGSLKDFSASELGA
MAV_1544|M.avium_104 -------------------MTTSVIVAGARTPIGKLMGSLKDFSASDLGA
MLBr_01158|M.leprae_Br4923 -------------------MTTSVIVTGARTPIGKLMGSLKDFSASDLGA
Mflv_2340|M.gilvum_PYR-GCK -------------------MTTSVIVAGARTPVGKLMGSLKEFSGSDLGA
MSMEG_4920|M.smegmatis_MC2_155 MRQLCRLIIVNTAIWGGSNMTTSVIVAGARTPVGKLMGSLKDFSGTDLGA
MAB_1463|M.abscessus_ATCC_1997 ----------------MAATNRSVIVAGARTPVGRFQGSLKDFSGAQLGG
Mvan_4891|M.vanbaalenii_PYR-1 ----------------MAADNSSVLVAGARTPIGKFMGSLAGLTGAELGG
TH_0255|M.thermoresistible__bu -------------------MRETVIVEAVRTAVGKRNGALADVHAADLSA
::* ..**.:*: *:* . .::*..
MMAR_4075|M.marinum_M IAIKAALEKGNVPASLVEYVIMGQVLTAG-AGQMPARQAAVAAGIGWDVP
MUL_3938|M.ulcerans_Agy99 IAIKAALEKGNVPASLVEYVIMGQVLTAG-AGQMPARQAAVAAGIGWDVT
Mb1358|M.bovis_AF2122/97 IAIKGALEKANVPASLVEYVIMGQVLTAG-AGQMPARQAAVAAGIGWDVP
Rv1323|M.tuberculosis_H37Rv IAIKGALEKANVPASLVEYVIMGQVLTAG-AGQMPARQAAVAAGIGWDVP
MAV_1544|M.avium_104 IAIAGALEKANVPADLVEYVIMGQVLTAG-AGQMPARQAAVAAGIGWDVP
MLBr_01158|M.leprae_Br4923 ITIAAALKKANVAPSIVQYVIIGQVLTAG-AGQMPARQAAVAAGIGWDVP
Mflv_2340|M.gilvum_PYR-GCK VAIKGALEKANVDASAVEYVIMGQVLSAG-AGQMPARQAAVAAGIPWDVA
MSMEG_4920|M.smegmatis_MC2_155 IAIRAALEKANVPASMVEYVIMGQVLTAG-AGQMPARQAAVAAGIPWDVA
MAB_1463|M.abscessus_ATCC_1997 IAIAGALAKAGVAPSAVDYVIMGQVLTAG-AGQMPARQAAVAGGIGMDVP
Mvan_4891|M.vanbaalenii_PYR-1 VAIAAALAKAGISGSQVQYVIMGQVLTAG-AGQIPARQAAVAGGIPMTVP
TH_0255|M.thermoresistible__bu VVLSELVERAGIDPELIDDVIWGCVSQVGDQSSNIGRYAVLAAGWPETIP
:.: : :..: . :: ** * * .* .. .* *.:*.* :.
MMAR_4075|M.marinum_M ALTINKMCLSGIDAIALADQLIRAGEFEVVVAGGQESMTKAPHLLMDSRS
MUL_3938|M.ulcerans_Agy99 ALTINKMCLSGIDAIALADQLIRAGEFEVVVAGGQESMTKAPHLLMDSRP
Mb1358|M.bovis_AF2122/97 ALTINKMCLSGIDAIALADQLIRAREFDVVVAGGQESMTKAPHLLMNSRS
Rv1323|M.tuberculosis_H37Rv ALTINKMCLSGIDAIALADQLIRAREFDVVVAGGQESMTKAPHLLMNSRS
MAV_1544|M.avium_104 ALSINKMCLSGIDAIALADQLIRAGEFDVVVAGGQESMTKAPHLLMDSRA
MLBr_01158|M.leprae_Br4923 ALTINKMCLSGLDAIALADQLIRAGEFDVVVAGGQESMTKAPHLLMDSRS
Mflv_2340|M.gilvum_PYR-GCK SLTINKMCLSGIDAIALADQLIRAGEFDVIVAGGQESMTQAPHLLMNSRS
MSMEG_4920|M.smegmatis_MC2_155 ALSINKMCLSGIDAIALADQLIRAGEFDVIVAGGQESMSQAPHLLPKSRE
MAB_1463|M.abscessus_ATCC_1997 ALTINKVCLSGVDAIALADQLIRAGEFEVVVAGGQESMTNAPHLLPKSRA
Mvan_4891|M.vanbaalenii_PYR-1 ALTVNKVCLSGLDAIALADQLIRAGAFDVVVAGGQESMSQAPHLLENLRA
TH_0255|M.thermoresistible__bu GTTVNRACGSSQQALDFAVHAVMSGQQDVVVAGGVEVMSRVP-LGAARAT
. ::*: * *. :*: :* : : : :*:**** * *:..* *
MMAR_4075|M.marinum_M GYKYGDVTVLDHMAYDGLHDAFTDQPMGALTEQRNDVDGFTRRQQDEYSA
MUL_3938|M.ulcerans_Agy99 GYKYGDVTVLDHMAYDGLHDAFTDQPMGALTEQRNDVDGFTRRQQDEYSA
Mb1358|M.bovis_AF2122/97 GYKYGDVTVLDHMAYDGLHDVFTDQPMGALTEQRNDVDMFTRSEQDEYAA
Rv1323|M.tuberculosis_H37Rv GYKYGDVTVLDHMAYDGLHDVFTDQPMGALTEQRNDVDMFTRSEQDEYAA
MAV_1544|M.avium_104 GYKYGDVTVLDHMAYDGLHDVFTDQPMGALTEQRNDVDKFTREEQDQYAA
MLBr_01158|M.leprae_Br4923 GYKYGDVTIVDHLAYDGLHDVFTNQPMGALTEQRNDVEKFTRQEQDEFAA
Mflv_2340|M.gilvum_PYR-GCK GYKYGDVTVLDHMAYDGLHDVFTDQPMGALTEQRNDADKFTRAEQDSYAA
MSMEG_4920|M.smegmatis_MC2_155 GYKYGDATLVDHLAYDGLHDVFTDQPMGALTEQRNDVDKFTRAEQDEYAA
MAB_1463|M.abscessus_ATCC_1997 GFKYGDVTLIDHMAFDGLHDAFTDQAMGLLTEQRNASDKFTREEQDEFAA
Mvan_4891|M.vanbaalenii_PYR-1 GTKFGDAALRDHVAVDGLRDVFTDQAMGSLTDS---VNALSRADQDEFAA
TH_0255|M.thermoresistible__bu GMPYGPKVLERYDNFG-----FNQGISAELIAER---WNLSRTRLDEYSA
* :* .: : . *.: . * . ::* *.::*
MMAR_4075|M.marinum_M ASHQKAAAAWKDGVFADEVVPVSVPQRKGDPLQFSEDEGIRANTTADSLA
MUL_3938|M.ulcerans_Agy99 ASHQKAAAAWKDGVFADEVVPVSVPQRKGDPLQFSEDEGIRANTTADSLA
Mb1358|M.bovis_AF2122/97 ASHQKAAAAWKDGVFADEVIPVNIPQRTGDPLQFTEDEGIRANTTAAALA
Rv1323|M.tuberculosis_H37Rv ASHQKAAAAWKDGVFADEVIPVNIPQRTGDPLQFTEDEGIRANTTAAALA
MAV_1544|M.avium_104 RSHQRAAAAWKDGVFTDEVVPVNIPQRKGDPLQFTEDEGIRANTTAESLA
MLBr_01158|M.leprae_Br4923 RSHQKAAAAWKDGVFADEVVPVSIPQSKGDSLQFTEDEGIRANTSAESLA
Mflv_2340|M.gilvum_PYR-GCK ESHQKAARAWKDGVYADEVVPVKIPQRKGDPIEFAEDEGIRADTTAESLG
MSMEG_4920|M.smegmatis_MC2_155 QSHQKAAAAWKDGVFADEVVPVSIPQRKGDPIEFAEDEGIRANTTAESLA
MAB_1463|M.abscessus_ATCC_1997 RSHQNAARGWKDGAFADEVVPVRIPQRKGDPIEFAEDEGIRADTTAASLA
Mvan_4891|M.vanbaalenii_PYR-1 ASHRRAAAARKNGVFDGEVVPVVIPQRKGDPVQVTVDEGIRADTTTASLA
TH_0255|M.thermoresistible__bu QSHERAAAAQSSGAFKDQIVPVFT-----DDAVVTEDEGIRRGTTVEKLA
**..** . ..*.: .:::** * .: ***** .*:. *.
MMAR_4075|M.marinum_M CLKPAFRKDGTVTAGSSSQISDGAAAVVVMSKEKAQELGLTWLVEIGAHG
MUL_3938|M.ulcerans_Agy99 GLKPAFRKDGTVTAGSSSQISDGAAAVVVMSKEKAQELGLTWLVEIGAHG
Mb1358|M.bovis_AF2122/97 GLKPAFRGDGTITAGSASQISDGAAAVVVMNQEKAQELGLTWLAEIGAHG
Rv1323|M.tuberculosis_H37Rv GLKPAFRGDGTITAGSASQISDGAAAVVVMNQEKAQELGLTWLAEIGAHG
MAV_1544|M.avium_104 GLKPAFRKDGTITAGSASQISDGAAAVVVMSKAKAQQLGLSWLAEIGAHG
MLBr_01158|M.leprae_Br4923 GLKPAFRCGGTITPGSASQISDGAATVVVMNKEKAQQLGLTWLVEIGAHG
Mflv_2340|M.gilvum_PYR-GCK GLRPAFRKDGTITAGSASQISDGACAVVVMNKAKAEELGLTWLCEIGAHG
MSMEG_4920|M.smegmatis_MC2_155 GLKPAFRKDGTITAGSASQISDGAAAVIVMNKAKAEELGLTWLAEIGAHG
MAB_1463|M.abscessus_ATCC_1997 GLRPAFSKDGTITAGSSSPISDGACAVVVMSKERAEREGLEWLAEIGAHG
Mvan_4891|M.vanbaalenii_PYR-1 RLRPAFVADGTVTAGNASTMNDGAAAVVVMSKAKATEVGLTWIAEIGAHG
TH_0255|M.thermoresistible__bu TLKPAFTENGVIHAGNSSQISDGAAALLVMTAETAVTLGLTPIVRYRAGA
*:*** .*.: .*.:* :.***.:::**. * ** : . * .
MMAR_4075|M.marinum_M VAAGPDSTLQSQPANAIKKALDKEGITADQLDVVEINEAFAAVALASTKE
MUL_3938|M.ulcerans_Agy99 VAAGPDSTLQSQPANAIKKALDKEGITADQLDVVEINEAFAAVALASTKE
Mb1358|M.bovis_AF2122/97 VVAGPDSTLQSQPANAINKALDREGISVDQLDVVEINEAFAAVALASIRE
Rv1323|M.tuberculosis_H37Rv VVAGPDSTLQSQPANAINKALDREGISVDQLDVVEINEAFAAVALASIRE
MAV_1544|M.avium_104 VVAGPDSTLQSQPANAIKKALGREGISVDQLDVVEINEAFAAVALASTRE
MLBr_01158|M.leprae_Br4923 VVAGPDSTLQSQPANAIKKAVDREGISVEQLDVVEINEAFAAVALASARE
Mflv_2340|M.gilvum_PYR-GCK VAAGPDSTLQSQPANAIRKALAKEGIGVDDLSVIEINEAFAAVALASARE
MSMEG_4920|M.smegmatis_MC2_155 VVAGPDSTLQSQPANAIKKAITREGITVDQLDVIEINEAFAAVALASTKE
MAB_1463|M.abscessus_ATCC_1997 VVAGPDSSLQLQPANAIRKACEREGISPEQLDLVEINEAFALVGLASIKD
Mvan_4891|M.vanbaalenii_PYR-1 VVAGPDSTLQMQPANAIAAACAIEGIDPAALDLVEINEAFAAVGIESTRA
TH_0255|M.thermoresistible__bu VTGADPVLMLTGPIPATEKVLRKTGLNIDDIGVFEVNEAFAPVPLAWQAE
*... : * * . *: :.:.*:***** * :
MMAR_4075|M.marinum_M LGLDPE----IVNVNGGAIAFGHPIGMSGARITLHVALELARRGSGYGVA
MUL_3938|M.ulcerans_Agy99 LGLDPE----IVNVNGGAIAFGHPIGMSGARITLHVALELARRGSGYGVA
Mb1358|M.bovis_AF2122/97 LGLNPQ----IVNVNGGAIAVGHPLGMSGTRITLHAALQLARRGSGVGVA
Rv1323|M.tuberculosis_H37Rv LGLNPQ----IVNVNGGAIAVGHPLGMSGTRITLHAALQLARRGSGVGVA
MAV_1544|M.avium_104 LGVNPD----IVNVNGGAIAVGHPIGMSGARITLHAALELSRRGSGYAVA
MLBr_01158|M.leprae_Br4923 LGIAPE----LVNVNGGAIAVGHPLGMSGARITLHVALELARRGSGYGVA
Mflv_2340|M.gilvum_PYR-GCK LGLDEATVAEKVNTNGGAIAIGHPIGMSGARITLHAALELARKGSGYAVA
MSMEG_4920|M.smegmatis_MC2_155 LGVDPA----KVNVNGGAIAIGHPIGMSGARIALHAALELARRGSGYAVA
MAB_1463|M.abscessus_ATCC_1997 LGIDPE----KVNVNGGAIAIGHPIGMSGARITLHLAQELKRRGGGIGVA
Mvan_4891|M.vanbaalenii_PYR-1 LGLDPE----RVNVNGGAIAIGHPLGMSGARIVLHLALELQRRGGGTGAA
TH_0255|M.thermoresistible__bu TGADPA----KLNPLGGAIALGHPLGASGAVLMTRMVHHMRANGIRYGLQ
* :* *****.***:* **: : : . .: .* .
MMAR_4075|M.marinum_M ALCGGGGQGDALILRAG-
MUL_3938|M.ulcerans_Agy99 ALCGGGGQGDALILRAG-
Mb1358|M.bovis_AF2122/97 ALCGAGGQGDALILRAG-
Rv1323|M.tuberculosis_H37Rv ALCGAGGQGDALILRAG-
MAV_1544|M.avium_104 ALCGAGGQGDALILRAG-
MLBr_01158|M.leprae_Br4923 ALCGAGGQGDALILRAV-
Mflv_2340|M.gilvum_PYR-GCK ALCGAGGQGDALILRRP-
MSMEG_4920|M.smegmatis_MC2_155 ALCGAGGQGDALVLRR--
MAB_1463|M.abscessus_ATCC_1997 ALCGGGGQGDALIVRV--
Mvan_4891|M.vanbaalenii_PYR-1 ALCGGGGQGSALIVRC--
TH_0255|M.thermoresistible__bu TMCEGGGTANATVVELLA
::* .** ..* ::.