For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSVKTRVLVIDDEPQILRALRINLSVRGYEVHTAASGAEALRMAADHKPDVVVLDLGLPDMTGIEVLAG LRGWLSAPVIVLSARSDSADKVEALDAGADDYVTKPFGMDEFLARLRAAVRRGATAAETDEPVIETSSFT VDLAAKKVTKGGVEVHLTPTEWGMLEMLVRHRGKLVGREELLKEVWGPAYAKETHYLRVYLAQLRRKLED DPSHPVHLLTEAGMGYRFQQ
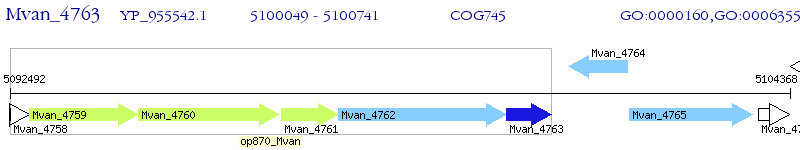
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4763 | - | - | 100% (230) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5009 | - | 5e-43 | 40.00% (225) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1752 | - | 1e-41 | 37.89% (227) | two component transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1055c | kdpE | 1e-106 | 83.93% (224) | transcriptional regulatory protein KdpE |
| M. gilvum PYR-GCK | Mflv_1744 | - | 4e-43 | 40.44% (225) | two component transcriptional regulator |
| M. tuberculosis H37Rv | Rv1027c | kdpE | 1e-106 | 83.93% (224) | transcriptional regulatory protein KdpE |
| M. leprae Br4923 | MLBr_02123 | - | 4e-43 | 41.18% (221) | two-component response regulator |
| M. abscessus ATCC 19977 | MAB_3250c | - | 3e-97 | 77.78% (225) | transcriptional regulatory protein KdpE |
| M. marinum M | MMAR_0635 | kdpE | 1e-110 | 88.34% (223) | transcriptional regulatory protein KdpE |
| M. avium 104 | MAV_1169 | - | 1e-110 | 86.16% (224) | KDP operon transcriptional regulatory protein KdpE |
| M. smegmatis MC2 155 | MSMEG_5396 | - | 1e-113 | 89.38% (226) | KDP operon transcriptional regulatory protein KdpE |
| M. thermoresistible (build 8) | TH_4129 | - | 1e-42 | 38.33% (227) | PUTATIVE DNA-binding response regulator MtrA |
| M. ulcerans Agy99 | MUL_0248 | prrA | 7e-43 | 40.72% (221) | two component response transcriptional regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1055c|M.bovis_AF2122/97 -----------MTLVLVIDDEPQILRALRINLTVRGYQVITASTGAGALR
Rv1027c|M.tuberculosis_H37Rv -----------MTLVLVIDDEPQILRALRINLTVRGYQVITASTGAGALR
MAV_1169|M.avium_104 -----------MTRVLVIDDEPQILRALRINLSVRGYEVVTASTGAGALR
Mvan_4763|M.vanbaalenii_PYR-1 ------MTS-VKTRVLVIDDEPQILRALRINLSVRGYEVHTAASGAEALR
MSMEG_5396|M.smegmatis_MC2_155 ------MTAPIKTRVLVIDDEPQILRALRINLSVRGYEVRTAASGAEALH
MMAR_0635|M.marinum_M -----------MTRVLVIDDEPQILRALRINFSVRGYDVVTAATGAAALR
MAB_3250c|M.abscessus_ATCC_199 -----------MTRVLVVDDEPQILRAMRINLSARGYEVVTASSGAGALR
MLBr_02123|M.leprae_Br4923 -----MDTGVSSPRVLVVDDDSDVLASLERGLRLSGFEVSTAIDGAEALR
MUL_0248|M.ulcerans_Agy99 --MGGMDTGANSPRVLVVDDDSDVLASLERGLRLSGFEVSTAVDGAEALR
Mflv_1744|M.gilvum_PYR-GCK -----MDSG--SPRVLVVDDDPDVLASLERGLRLSGFDVFTAVDGAEALR
TH_4129|M.thermoresistible__bu LSRFGDNMDTMRQRILVVDDDPSLAEMLTIVLRGEGFDTAVIGDGSQALT
:**:**:..: : : *::. . *: **
Mb1055c|M.bovis_AF2122/97 AAAEHPPDVVILDLGLPDMSGIDVLGGLRG-WLTAPVIVLSARTDSSDKV
Rv1027c|M.tuberculosis_H37Rv AAAEHPPDVVILDLGLPDMSGIDVLGGLRG-WLTAPVIVLSARTDSSDKV
MAV_1169|M.avium_104 AAAEHKPDVVILDLGLPDISGIDVLAGLRG-WLTAPVIVLSARTDSSDKV
Mvan_4763|M.vanbaalenii_PYR-1 MAADHKPDVVVLDLGLPDMTGIEVLAGLRG-WLSAPVIVLSARSDSADKV
MSMEG_5396|M.smegmatis_MC2_155 SAADHRPDVIVLDLGLPDMSGIEVLHGLRG-WLSAPVIVLSARTDSSDKV
MMAR_0635|M.marinum_M AAAEQRPDVVILDLGLPDMSGIEVLAGLRG-WFSAPVIVLSARSDSSDKV
MAB_3250c|M.abscessus_ATCC_199 AAAETQPEVVILDLGLPDMDGTEVLAGLRG-WTDVPVIVLSARTDSADKV
MLBr_02123|M.leprae_Br4923 NATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVLSARSSVDDRV
MUL_0248|M.ulcerans_Agy99 SATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVLSARSSVDDRV
Mflv_1744|M.gilvum_PYR-GCK SATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVLSARSSVDDRV
TH_4129|M.thermoresistible__bu AVRELRPDLVLLDLMLPGMNGIDVCRVLRA-DSGVPIVMLTAKSDTVDVV
. : *: ::**: :* : * .* **. .*: :*:*::. * *
Mb1055c|M.bovis_AF2122/97 QALDAGADDYVTKPFGMDEFLARLRAAVRRNTAAAELEQPVIETDSFTVD
Rv1027c|M.tuberculosis_H37Rv QALDAGADDYVTKPFGMDEFLARLRAAVRRNTAAAELEQPVIETDSFTVD
MAV_1169|M.avium_104 EALDAGADDYVTKPFGMDEFLARLRAAVRRNTAASEMEQPVVETESFTVD
Mvan_4763|M.vanbaalenii_PYR-1 EALDAGADDYVTKPFGMDEFLARLRAAVRRGATAAETDEPVIETSSFTVD
MSMEG_5396|M.smegmatis_MC2_155 EALDAGADDYVTKPFGMDEFLARLRAAARR-AAVPETDQPVIETTSFTVD
MMAR_0635|M.marinum_M QALDAGADDYVTKPFGMDELLARLRAAVRRAATATDTDQPVIETSSFTVD
MAB_3250c|M.abscessus_ATCC_199 EALDAGADDYVTKPFGMDEFLARLRAAVRR--GTPDSAEPVVHTETFDVD
MLBr_02123|M.leprae_Br4923 AGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSETIAVGPLEVD
MUL_0248|M.ulcerans_Agy99 AGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSEIITVGPLEVD
Mflv_1744|M.gilvum_PYR-GCK AGLEAGADDYLVKPFVLAELVARVKALLRRRGSTATFSSETITVGPLEVD
TH_4129|M.thermoresistible__bu LGLESGADDYVVKPFKPKELVARVRARLRR---NEDEPAEMLSIGDIDID
.*::*****:.*** *::**::* ** : : :*
Mb1055c|M.bovis_AF2122/97 LAGKKVIKDGAEVHLTPTEWGMLEMLARNRGKLVGRGELLKEVWGPAYAT
Rv1027c|M.tuberculosis_H37Rv LAGKKVIKDGAEVHLTPTEWGMLEMLARNRGKLVGRGELLKEVWGPAYAT
MAV_1169|M.avium_104 LAAKKVTKNGSEVHLTPTEWGMLEVLVRNRGKLVGREELLKEVWGPAYAT
Mvan_4763|M.vanbaalenii_PYR-1 LAAKKVTKGGVEVHLTPTEWGMLEMLVRHRGKLVGREELLKEVWGPAYAK
MSMEG_5396|M.smegmatis_MC2_155 LAAKKVIKNGAEVHLTPTEWGMLEMLVRNRGKLVGREELLREVWGPAYAT
MMAR_0635|M.marinum_M LATKKVIKNGSEVHLTPTEWGMLEMLVRNRGKLVGREELLKEVWGPAYAK
MAB_3250c|M.abscessus_ATCC_199 LSSHKVIRDGREVHLTPTEWSMLAVLVRNRGKLVGQKELLHEVWGPAYSS
MLBr_02123|M.leprae_Br4923 IPGRRARVNGVDVDLTKREFDLLAVLAEHKTTVLSRAQLLELVWGYDFAA
MUL_0248|M.ulcerans_Agy99 IPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQLLELVWGYDFAA
Mflv_1744|M.gilvum_PYR-GCK IPGRRARVDGVDVDLTKREFDLLAVLAEHKTAVLSRAQLLELVWGYDFAA
TH_4129|M.thermoresistible__bu VPAHKVTRNGEQIQLTPLEFDLLVALARKPRQVFTRDVLLEQVWGYRHPA
:. ::. .* ::.** *:.:* *..: :. : **. *** ..
Mb1055c|M.bovis_AF2122/97 ETHYLRVYLAQLRRKLEDDPSHPKHLLTESGMGYRFEA-
Rv1027c|M.tuberculosis_H37Rv ETHYLRVYLAQLRRKLEDDPSHPKHLLTESGMGYRFEA-
MAV_1169|M.avium_104 ETHYLRVYLAQLRRKLENDPSHPKHLLTESGMGYRFEA-
Mvan_4763|M.vanbaalenii_PYR-1 ETHYLRVYLAQLRRKLEDDPSHPVHLLTEAGMGYRFQQ-
MSMEG_5396|M.smegmatis_MC2_155 ETHYLRVYLAQLRRKLEDDPSHPVHLLTEAGMGYRFEA-
MMAR_0635|M.marinum_M ETHYLRVYLAQLRRKLEDDPAHPRHLLTEAGMGYRFDG-
MAB_3250c|M.abscessus_ATCC_199 ETHYLRVYLAQLRRKLEIDPAHPKHLLTEAGMGYRFQE-
MLBr_02123|M.leprae_Br4923 DTNVVDVFIGYLRRKLEAN-SGPRLLHTVRGVGFVLRMQ
MUL_0248|M.ulcerans_Agy99 DTNVVDVFIGYLRRKLEAN-GGPRLLHTVRGVGFVLRMH
Mflv_1744|M.gilvum_PYR-GCK DTNVVDVFIGYLRRKLEAN-GAPRLLHTVRGVGFVLRQQ
TH_4129|M.thermoresistible__bu DTRLVNVHVQRLRAKVEKDPENPQVVLTVRGVGYKAGPP
:*. : *.: ** *:* : * : * *:*: