For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSVDDVVSGTADDSRKQNRYYFDRHTPEYRLQFEKITEEMQTKCPMAWSDTYGGHWVAADSKHVFELARC PVVSNHHDISGETPFQGITIPKASRATVVRGGILEMDEPEHSAYRGALNPYLSPAAIKRWEPFIHEITRA ALDEHIESGRIDFVDHLANVVPAVFTLAMMGIKLDKWNVYSEPTHASVYTPEHAPEREKINEQHRAMGLD LITNMMEIRENPRPGLVNALLQLRIDGEPAPDIEILGNLGLIIGGGFDTTTALTAHALEWLGEHPDQRQR LSSERATLLDPATEEFLRFFTPAPGDGRTFADDVEVEGQRFKKFERLWLSWAMANRDASVFDRPNEVVLD RKGNRHFSFGIGVHRCVGSNVARTLFKSMVTAVLDRMPDYVCDPEGTVHYDTIGVIQGMRNLPATFTPGK RLGPGLDETLEKLQRICDEQELARPITERKEAAVID
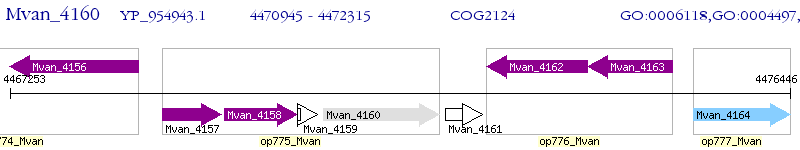
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4160 | - | - | 100% (456) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_4880 | - | e-179 | 64.94% (445) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_4979 | - | 6e-44 | 31.99% (397) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0789c | cyp123 | 1e-29 | 29.17% (360) | cytochrome P450 123 |
| M. gilvum PYR-GCK | Mflv_2498 | - | 0.0 | 92.11% (456) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv0766c | cyp123 | 1e-29 | 29.17% (360) | cytochrome P450 123 |
| M. leprae Br4923 | MLBr_02088 | - | 6e-23 | 25.47% (318) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_3825 | - | 3e-29 | 27.19% (320) | cytochrome P450 |
| M. marinum M | MMAR_4717 | cyp188A3 | 0.0 | 74.62% (457) | cytochrome P450 188A3 Cyp188A3 |
| M. avium 104 | MAV_0940 | - | 0.0 | 76.59% (457) | cytochrome P450 superfamily protein |
| M. smegmatis MC2 155 | MSMEG_4803 | - | 0.0 | 87.28% (456) | cytochrome P450 superfamily protein |
| M. thermoresistible (build 8) | TH_2693 | - | 0.0 | 87.47% (455) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0317 | cyp188A3 | 1e-177 | 73.05% (397) | cytochrome P450 188A3 Cyp188A3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4160|M.vanbaalenii_PYR-1 -----------------MSVDDVVSGTADDSRKQNRYYFDRHTPEYRLQF
Mflv_2498|M.gilvum_PYR-GCK --MTNYTEIAASKESRLLSVDDVVDGTADDSRRKNTYYFDRHTPEYRLQF
MSMEG_4803|M.smegmatis_MC2_155 -----------------MSIQDVTT---DDDREKITYHFDRHTPEYRLQF
TH_2693|M.thermoresistible__bu -----------------LSVDDIIT---DAERKKHRYHFDRHTPEYRLQF
MMAR_4717|M.marinum_M -----------------MSVDDVVS---DSDHQKNRYQFDRHSPEYRSQF
MUL_0317|M.ulcerans_Agy99 -----------------MSVDDVVS---DSDHHQNRYQFDRHSPEYRSQF
MAV_0940|M.avium_104 -----------------MSVDDGVN---DSDRKKNRYHFDRHSPDYRSRF
Mb0789c|M.bovis_AF2122/97 ---------------------------MTVRVGDPELVLDPYDYDFHEDP
Rv0766c|M.tuberculosis_H37Rv ---------------------------MTVRVGDPELVLDPYDYDFHEDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
MAB_3825|M.abscessus_ATCC_1997 -----------------------MATALSEIEEAGRVLADPAAYADELRL
.
Mvan_4160|M.vanbaalenii_PYR-1 EKITEEMQTKCPMA--WSDTYGGHWVAADSKHVFELARCPVVSNHHDISG
Mflv_2498|M.gilvum_PYR-GCK DKITEEMHAKCPMA--WSETYNGHWVAADSKHVFELARCPAVSNHHDISG
MSMEG_4803|M.smegmatis_MC2_155 EKITEEMHSRCPVA--WTETYDGHWVAAGSNEVFELARCPVVSNHHDISG
TH_2693|M.thermoresistible__bu EKITEEMHAKCPMA--WTDTYGGHWVAADSKHVFELARCPVVSNHHDITG
MMAR_4717|M.marinum_M KAITEEMHTTCPMA--WSDTYGGHWVAAGSHEVFELARCPAVSNDHDVNN
MUL_0317|M.ulcerans_Agy99 KAITEEMHTTCPMA--WSDTYGGHWVAAGSHEVFELARCPAVSNDHDVNN
MAV_0940|M.avium_104 KAITEEMHAKCPMA--WTDTYGGHWVAAGSHEVFELARCPAVSNDHDING
Mb0789c|M.bovis_AF2122/97 YPYYRRLRDEAPLY--RNEERNFWAVSRHHDVLQGFRDSTALSNAYGVSL
Rv0766c|M.tuberculosis_H37Rv YPYYRRLRDEAPLY--RNEERNFWAVSRHHDVLQGFRDSTALSNAYGVSL
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQ--LPG-MPLTVFSSFSDCDEALRHPLSASDRLKATL
MAB_3825|M.abscessus_ATCC_1997 HAAMTLLRREQPVTKVITDDYRPFWAVTKHDDIMAVERDNALWINEPRPL
: *: . .
Mvan_4160|M.vanbaalenii_PYR-1 ETP-FQGITIPKASRATVVRGGILEMDEPEHSAYRGALNPYLSPAAIKRW
Mflv_2498|M.gilvum_PYR-GCK ETP-FQGITIPKASRATVVRGGILEMDEPEHSAYRGALNPYLSPAAIKRW
MSMEG_4803|M.smegmatis_MC2_155 ETP-FQGITIPKASRATVVRGGILEMDEPEHSAYRGALNPYLSPAAIKRW
TH_2693|M.thermoresistible__bu ETP-FQGITIPKASRATVVRGGILEMDEPEHSIYRGALNPYLSPAAIKRW
MMAR_4717|M.marinum_M QRRGYKGISIPTASRINGVRGGILEMDDPEHRIYRTVLNPYLSPAAVKRW
MUL_0317|M.ulcerans_Agy99 QRRGYKGISIPTASRINGVRGGILEMDDPEHRIYRTVLNPYLSPAAVKRW
MAV_0940|M.avium_104 ERRGYKGISIPTASRVSAVRGGILEMDDPEHRIYRTVLNPYLSPAAVKRW
Mb0789c|M.bovis_AF2122/97 DPSSRTS----EAYRVMS----MLAMDDPAHLRMRTLVSKGFTPRRIREL
Rv0766c|M.tuberculosis_H37Rv DPSSRTS----EAYRVMS----MLAMDDPAHLRMRTLVSKGFTPRRIREL
MLBr_02088|M.leprae_Br4923 AQQAIAAGAEPRPFYASS----FMFLDPPDHTRLRKLVSKAFAPKVVQAL
MAB_3825|M.abscessus_ATCC_1997 LMNLEQEAELDKQAAMGIELKTLVHIDDPKHRVLRAIGADWFRPKAMRDM
:: :* * * * : * ::
Mvan_4160|M.vanbaalenii_PYR-1 EPFIHEITRAALDEHIESG-RIDFVDHLANVVPAVFTLAMMGIKLDKWNV
Mflv_2498|M.gilvum_PYR-GCK EPFIDEITRAALDEHIESG-RIDFVDHLANVVPAVFTLAMMGIELKKWNV
MSMEG_4803|M.smegmatis_MC2_155 EPFVDEITRAAIDEHISSG-RIDFVEHLANVVPAVFTLAMMGIELKKWNV
TH_2693|M.thermoresistible__bu EPFVHEITRAAIDEKIESG-EIDFVEDLANVVPAVFTLAMMGIELKKWKV
MMAR_4717|M.marinum_M EPFIDEVTRACLDERIEQG-SIDFVDDLANIVPAVLTLAILGIPLEKWNL
MUL_0317|M.ulcerans_Agy99 EPFIDEVTRACLDERIEQG-SIDFVDDLANIVPAVLTLAILGIPLEKWNL
MAV_0940|M.avium_104 EPFIDEVTRAALDEKIEEG-SIDFVDDLANIVPAVLTLAMLGIPLKKWKM
Mb0789c|M.bovis_AF2122/97 EPQVLELARIHLDSALQTE-SFDFVAEFAGKLPMDVISELIGVPDTDRAR
Rv0766c|M.tuberculosis_H37Rv EPQVLELARIHLDSALQTE-SFDFVAEFAGKLPMDVISELIGVPDTDRAR
MLBr_02088|M.leprae_Br4923 EGDIAALVDSLLDKGAAAG-QFDVIADLAFPLAVAVICRLLGVPYEDAPE
MAB_3825|M.abscessus_ATCC_1997 KLRTDELANRYVNKLLEAGGSCDFAQDIAVHFPLYVIMSLLGIPESDFDR
: :. ::. *. .:* .. . ::*: .
Mvan_4160|M.vanbaalenii_PYR-1 YSEPTHASVYTPEH----APEREKINEQHRAMGLDLITN----MMEIREN
Mflv_2498|M.gilvum_PYR-GCK YSEPTHASVYTPEH----APEREKINEQHREMGIDLINN----MMEIREN
MSMEG_4803|M.smegmatis_MC2_155 YSEPTHASVYTPEH----APEREKINEQHREMGIDLINN----MMEIRQN
TH_2693|M.thermoresistible__bu YSEPTHASVYTPEH----APEREKVNEMHREMGIDLINN----MMEIREN
MMAR_4717|M.marinum_M YSEPVHAAVYTPEH----SPDIQRVTEMHRQMGLDMVTN----MIEIRDN
MUL_0317|M.ulcerans_Agy99 YSEPVHAAVYTPEH----SPDIQRVTEMHRQMGLDMVTN----MIEIRDN
MAV_0940|M.avium_104 YSEPVHAAVYTPEH----SPDIERVTAMHREMGLDMVNN----MLEIREN
Mb0789c|M.bovis_AF2122/97 IRALADAVLHRED---GVADVPP----PAMAASIELMRYYADLIAEFRRR
Rv0766c|M.tuberculosis_H37Rv IRALADAVLHRED---GVADVPP----PAMAASIELMRYYADLIAEFRRR
MLBr_02088|M.leprae_Br4923 FGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGT
MAB_3825|M.abscessus_ATCC_1997 MLKLTQELFGGDDS----EFQRGTTPEEQLMALLDFFGYFSGLTASRRQH
. : : : *
Mvan_4160|M.vanbaalenii_PYR-1 PRPGLVNALLQLRIDGEPAPDIEILGNLGLIIGGGFDTTTALTAHALEWL
Mflv_2498|M.gilvum_PYR-GCK PRPGLVNALLQLRIDGEPAPDMEILGNLGLIIGGGFDTTTALTAHALEWL
MSMEG_4803|M.smegmatis_MC2_155 PRPGLVNALLQLRIDGEPAPDMEILGNLGLIIGGGFDTTTALTAHALEWL
TH_2693|M.thermoresistible__bu PRPGLVNALLQLRIDGEPAPDMEILGNLGLIIGGGFDTTTALTAHALEWL
MMAR_4717|M.marinum_M PRPGLVNALVQMRIDGQPAPDLEILGNLGLVIGGGFDTTTALTAHALEWL
MUL_0317|M.ulcerans_Agy99 PRPGLVNALVQMRIDGQPAPDLEILGNLGLVIGGGFDTTTALTAHALEWL
MAV_0940|M.avium_104 PRPGIVNALLQMRIDGEPAPDLEILGNLGLVIGGGFDTTTALTAHSLEWL
Mb0789c|M.bovis_AF2122/97 PANNLTSALLAAELDGDRLSDQEIMAFLFLMVIAGNETTTKLLANAVYWA
Rv0766c|M.tuberculosis_H37Rv PANNLTSALLAAELDGDRLSDQEIMAFLFLMVIAGNETTTKLLANAVYWA
MLBr_02088|M.leprae_Br4923 PGEDLISRLIELDESGDQLTEEEIIATCGLLLVAGHETTVNLIANAVLAM
MAB_3825|M.abscessus_ATCC_1997 PTDDLASTIANARVDGELLSDVDTASYYTIIATAGHDTTSATIAGGLEAL
* .: . : .*: .: : . :: .* :** * .:
Mvan_4160|M.vanbaalenii_PYR-1 GEHPDQRQRLSSERATLLDPATEEFLRFFTPAPGDGRTFADDVEVEGQRF
Mflv_2498|M.gilvum_PYR-GCK GEHPDERERLSRERATLLAPATEEFLRFFTPAPGDGRTFSEDVEVAGQRF
MSMEG_4803|M.smegmatis_MC2_155 GEHPDERERLSRERDTLLNPATEEFLRFFTPAPGDGRTFAEDAEVAGYKF
TH_2693|M.thermoresistible__bu GEHPDERERLSRERDTLLNPATEEFLRFFTPAPGDGRTFAEDVEVLGTQF
MMAR_4717|M.marinum_M SEHPDERERLWRNRDTLIDSATEEFLRYFTPAPGDGRTFSDDVEFDGIHF
MUL_0317|M.ulcerans_Agy99 SEHHDERERLWRNRDTLIDSATEEFLRYFTPAPGDGRTFSDDLEFFGIHF
MAV_0940|M.avium_104 SEHPEQRQLLSDERKTLLDPATEEFLRYFTPAPGDGRTFSEDFELDGTVF
Mb0789c|M.bovis_AF2122/97 AHHPGQLARVFAD-HSRIPMWVEETLRYDTSSQILARTVAHDLTLYDTTI
Rv0766c|M.tuberculosis_H37Rv AHHPGQLARVFAD-HSRIPMWVEETLRYDTSSQILARTVAHDLTLYDTTI
MLBr_02088|M.leprae_Br4923 LRNPSQWKALSSN-PQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQTTL
MAB_3825|M.abscessus_ATCC_1997 LEHPDQLARLREN-PGLMPLAVDEMIRWVTPVKEFMRTATADTEIRGVPI
.: : : : .:* :*: .. *. : * . :
Mvan_4160|M.vanbaalenii_PYR-1 KKFERLWLSWAMANRDASVFDRPNEVVLDRKG-NRHFSFGIGVHRCVGSN
Mflv_2498|M.gilvum_PYR-GCK KQYERLWLSWAMANRDPAVFDEPNKVVLDRKG-NRHFSFGIGVHRCVGSN
MSMEG_4803|M.smegmatis_MC2_155 KQYERLWLSWAMANRDPSVFDKPNEVILDRKG-NRHFSFGIGVHRCVGSN
TH_2693|M.thermoresistible__bu KKYERLWLSWAMANRDPAVFERPNEIILDRKG-NRHFSFGIGVHRCVGSN
MMAR_4717|M.marinum_M KEGERLWISWAMANRDPQVFPDPDELVLDRRG-NRHFSFGIGVHRCIGSN
MUL_0317|M.ulcerans_Agy99 KEGERLWISWAMANREPQVFPDPDELVLDRRG-NRHFSFGIGVHRCVGSN
MAV_0940|M.avium_104 KEGERLWISWAMANRDPAVFHDPDEVILDRKG-NRHFSFGLGIHRCIGSN
Mb0789c|M.bovis_AF2122/97 PEGEVLLLLPGSANRDDRVFDDPDDYRIGREIGCKLVSFGSGAHFCLGAH
Rv0766c|M.tuberculosis_H37Rv PEGEVLLLLPGSANRDDRVFDDPDDYRIGREIGCKLVSFGSGAHFCLGAH
MLBr_02088|M.leprae_Br4923 TEGDTMVLLLAAANRDPAVYSRPDEFDPDRPS-SRHLAFAVGSHFCLGAA
MAB_3825|M.abscessus_ATCC_1997 TEGESVLLSYPSGNRDEDIFTDPFTFDIARDP-NKHLAFGFGVHFCLGAA
: : : : .**: :: * * : .:*. * * *:*:
Mvan_4160|M.vanbaalenii_PYR-1 VARTLFKSMVTAVLDRMPDYVCDPEGTVHYDTIGVIQGMRNLPATFTPGK
Mflv_2498|M.gilvum_PYR-GCK VARTVFKSMLTAVLDRMPDYVCDPEGTVHYDSIGVIQGMRNLPATFTPGK
MSMEG_4803|M.smegmatis_MC2_155 VARTVFKSMLTAVLDRMPDYVCDPEGTVHYDTIGVIQGMRNLPATFTPSR
TH_2693|M.thermoresistible__bu VARTVFKSMLTAVLDRMPDYVCDPEGTMHYDTIGVIQGMRHLPAKFTPGK
MMAR_4717|M.marinum_M VARTVFKSMLSAVADRMPDYRCDPDGAVHYETIGVIQGMRKLPATFTPGR
MUL_0317|M.ulcerans_Agy99 VARTVFKSMLSAVADRMPDYRCDPDGRSTTRPSASSRACASCPQRSPRAA
MAV_0940|M.avium_104 VARTVFKSMLIAVLDRMPDYRCDPEGTVHYETIGVIQGMRKLPATFTPGR
Mb0789c|M.bovis_AF2122/97 LARMEARVALGALLRRIRNYEVDDD-NVVRVHSSNVRGFAHLPISVQAR-
Rv0766c|M.tuberculosis_H37Rv LARMEARVALGALLRRIRNYEVDDD-NVVRVHSSNVRGFAHLPISVQAR-
MLBr_02088|M.leprae_Br4923 LARLEATVTLSAISARFPQVQLAGE-LVYKPN-VAMRGMSALPVQV----
MAB_3825|M.abscessus_ATCC_1997 LARMEVNSFFSALLPRLESIEVDGN--IERTSTIFVGGIKHLPIRYTLR-
:** . *: *: . : . *
Mvan_4160|M.vanbaalenii_PYR-1 RLGPGLDETLEKLQRICDEQELAR-------------------PITERKE
Mflv_2498|M.gilvum_PYR-GCK PLGPGLDETIEKLQRICNEQELAR-------------------PITERKE
MSMEG_4803|M.smegmatis_MC2_155 PLGPGLDETLEKLQRICNEEELAR-------------------PITERKE
TH_2693|M.thermoresistible__bu PLGPKLDETLDRLQQICDEQELAR-------------------PITERKE
MMAR_4717|M.marinum_M KAGAGLDETLAKLQRICDEQELAR-------------------PVTERKQ
MUL_0317|M.ulcerans_Agy99 RSAR----AWTRLSRSCNASATNRNSRGPSPSANKPRSSTDAPPASRHKI
MAV_0940|M.avium_104 RIGAGLDETLEKLQRICDEQELAR-------------------PITERKE
Mb0789c|M.bovis_AF2122/97 --------------------------------------------------
Rv0766c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02088|M.leprae_Br4923 --------------------------------------------------
MAB_3825|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_4160|M.vanbaalenii_PYR-1 AAVID-
Mflv_2498|M.gilvum_PYR-GCK AAVID-
MSMEG_4803|M.smegmatis_MC2_155 RAVID-
TH_2693|M.thermoresistible__bu PAVITW
MMAR_4717|M.marinum_M AAVID-
MUL_0317|M.ulcerans_Agy99 ASGSGR
MAV_0940|M.avium_104 AAVID-
Mb0789c|M.bovis_AF2122/97 ------
Rv0766c|M.tuberculosis_H37Rv ------
MLBr_02088|M.leprae_Br4923 ------
MAB_3825|M.abscessus_ATCC_1997 ------