For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEIRDAADPAPNPHATAEEVEAAMHDSKLAQVLYHDWEAETYDEKWSISYDQRCIDYARGRFDAIVPED EQRKLPYDRALELGCGTGFFLLNLIQSGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYE DDTFDLVVGHAVLHHIPDVELSLREVVRILKPGGRFVFAGEPTTVGNRYARELSTLTWHATTNLTKLPFL SGWRRPQEELDESSRAAALEAVVDLHTFDPADLERMATNAGAVEVRTASEEFTAAMLGWPVRTFEAAVPP GRLGWGWAKFAFNSWTTLSWVDANVWRKVVPKGWFYNVMVTGVKPS
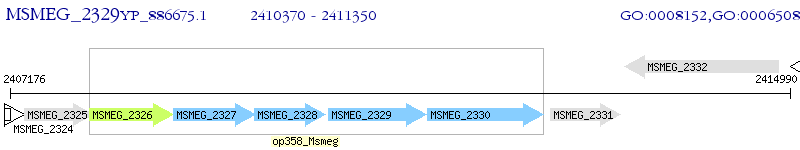
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2329 | - | - | 100% (326) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_1049 | - | 0.0 | 100.00% (326) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_6483 | - | 2e-10 | 31.48% (108) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3064c | - | 1e-165 | 85.89% (319) | hypothetical protein Mb3064c |
| M. gilvum PYR-GCK | Mflv_4269 | - | 1e-178 | 91.90% (321) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3038c | - | 1e-165 | 85.89% (319) | hypothetical protein Rv3038c |
| M. leprae Br4923 | MLBr_01723 | - | 1e-154 | 78.46% (325) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_3381c | - | 1e-161 | 82.52% (326) | hypothetical protein MAB_3381c |
| M. marinum M | MMAR_1663 | - | 1e-167 | 86.52% (319) | hypothetical protein MMAR_1663 |
| M. avium 104 | MAV_3903 | - | 1e-170 | 87.81% (320) | methyltransferase, UbiE/COQ5 family protein |
| M. thermoresistible (build 8) | TH_2189 | - | 1e-170 | 88.01% (317) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1900 | - | 1e-166 | 86.21% (319) | hypothetical protein MUL_1900 |
| M. vanbaalenii PYR-1 | Mvan_2090 | - | 1e-179 | 92.21% (321) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1663|M.marinum_M --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4269|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2090|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu VIRRLMATALTALTTAVLAGCGDAGSWVEAKPAEGWSAQYGDAANSSYTS
MAV_3903|M.avium_104 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1663|M.marinum_M --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4269|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2090|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu AAGAEALTLEWSRSVKGELAAQVAVGASGYLAVNAQTPAGCSLMVWEYAN
MAV_3903|M.avium_104 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1663|M.marinum_M --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4269|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2090|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu SARQRWCTRLVQGGGRTSPLLDGFDNVYIGQPGAILSFPPTQWIRWRKPV
MAV_3903|M.avium_104 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 --------------------------------------------------
Rv3038c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1663|M.marinum_M --------------------------------------------------
MUL_1900|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4269|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2090|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2329|M.smegmatis_MC2_155 --------------------------------------------------
TH_2189|M.thermoresistible__bu IGMPTTPRILAPGELLVVTHLGQVLLFDAHRGTVTGTPLDLVAGVDPTDS
MAV_3903|M.avium_104 --------------------------------------------------
MAB_3381c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
Mb3064c|M.bovis_AF2122/97 ------------------------------------------MTRSSN--
Rv3038c|M.tuberculosis_H37Rv ------------------------------------------MTRSSN--
MMAR_1663|M.marinum_M ------------------------------------------MTRSSE--
MUL_1900|M.ulcerans_Agy99 ------------------------------------------MTRSSE--
Mflv_4269|M.gilvum_PYR-GCK ------------------------------------------MTETKEPG
Mvan_2090|M.vanbaalenii_PYR-1 ------------------------------------------MTETKEPG
MSMEG_2329|M.smegmatis_MC2_155 ------------------------------------------MTEIR---
TH_2189|M.thermoresistible__bu ERGLADCAGARXXXCWPPPRPPSTVTPRARDAARHPTARLPAMTETTEPG
MAV_3903|M.avium_104 ------------------------------------------MTRST---
MAB_3381c|M.abscessus_ATCC_199 ------------------------------------------MTEITG--
MLBr_01723|M.leprae_Br4923 ------------------------------------------MTRSTE--
**.
Mb3064c|M.bovis_AF2122/97 ---IPADATPN---------PHATAEQVAAARHDSKLAQVLYHDWEAENY
Rv3038c|M.tuberculosis_H37Rv ---IPADATPN---------PHATAEQVAAARHDSKLAQVLYHDWEAENY
MMAR_1663|M.marinum_M ---IPADAAPN---------PHATAEEVEAARHDSKLAQVLYHDWEAENY
MUL_1900|M.ulcerans_Agy99 ---IPADAAPN---------PHATAEEVEAARHDSKLAQVLYHDWEAENY
Mflv_4269|M.gilvum_PYR-GCK AYPDVAVPAPT---------PHATKEQVEAALHDTKLAQVLYHDWEAETY
Mvan_2090|M.vanbaalenii_PYR-1 TYPDVAVPAPTPDVAVPAPTPRATKEQVEAAMHDTKLAQVLYHDWEAETY
MSMEG_2329|M.smegmatis_MC2_155 ---DAADPAPN---------PHATAEEVEAAMHDSKLAQVLYHDWEAETY
TH_2189|M.thermoresistible__bu IGSEGAGDLPAPN-------PHATAEQVEAAMRDSKLAQILYHDWEAESY
MAV_3903|M.avium_104 ------DAVPT---------PHATAEQVEAARHDSKLAQVLYHDWEAETY
MAB_3381c|M.abscessus_ATCC_199 -----LDGQPT---------TKATAEQVAAAFEDNKLAQILYHDWEAETY
MLBr_01723|M.leprae_Br4923 ---ISANAVPN---------PHATAEQVAAARKDSKLAQVLYHDWEAENY
* .:** *:* ** .*.****:********.*
Mb3064c|M.bovis_AF2122/97 DEKWSISYDQRCVDYARGRFDAIVPDEVIAQLPYDRALELGCGTGFFLLN
Rv3038c|M.tuberculosis_H37Rv DEKWSISYDQRCVDYARGRFDAIVPDEVIAQLPYDRALELGCGTGFFLLN
MMAR_1663|M.marinum_M DEKWSISYDQRCVEYARGRFDAIVPAQVHADLPYERALELGCGTGFFLLN
MUL_1900|M.ulcerans_Agy99 DEKWSISYDQRCVEYARGRFDAIVPAQVHADLPYERALELGCGTGFFLLN
Mflv_4269|M.gilvum_PYR-GCK DDKWSISYDQRCVDYARGRFDAAVPEEVQRELPYDRALELGCGTGFFLLN
Mvan_2090|M.vanbaalenii_PYR-1 DEKWSISYDQRCVDYARGRFDAAVPEEVQRELPYDRALELGCGTGFFLLN
MSMEG_2329|M.smegmatis_MC2_155 DEKWSISYDQRCIDYARGRFDAIVPEDEQRKLPYDRALELGCGTGFFLLN
TH_2189|M.thermoresistible__bu DDKWSISYDQRCVDYARGRFDAIVPDHVQRELPYDNALELGCGTGFFLLN
MAV_3903|M.avium_104 DEKWSISYDQRCIDYARGRFDAIVPEHVLRELPYDRALELGCGTGFFLLN
MAB_3381c|M.abscessus_ATCC_199 DEKWSISYDQRCIDYARGRFDAIVPVEDQRELPYDRALELGCGTGFFLLN
MLBr_01723|M.leprae_Br4923 DEKWSISYDQRCVDYVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLN
*:**********::*.* *** ** . .***: **************
Mb3064c|M.bovis_AF2122/97 LIQAGVARRGSVTDLSPGMVKVATRNGQALGLDIDGRVADAEGIPYDDDA
Rv3038c|M.tuberculosis_H37Rv LIQAGVARRGSVTDLSPGMVKVATRNGQALGLDIDGRVADAEGIPYDDDA
MMAR_1663|M.marinum_M LIQAGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYDDDT
MUL_1900|M.ulcerans_Agy99 LIQAGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYDDDT
Mflv_4269|M.gilvum_PYR-GCK LIQAGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYEDNT
Mvan_2090|M.vanbaalenii_PYR-1 LIQAGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYEDNT
MSMEG_2329|M.smegmatis_MC2_155 LIQSGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYEDDT
TH_2189|M.thermoresistible__bu LIQSGVARRGSVTDLSPGMVRVATRNGRSLGLDVDGRVADAEGIPYDDNT
MAV_3903|M.avium_104 LIQSGVARRGSVTDLSPGMVKVATRNGQSLGLDIDGRVADAEGIPYEDDT
MAB_3381c|M.abscessus_ATCC_199 LMQAGVARRGSVTDLSPGMVKVATRTGQELGLDVDGRVADAERIPYDDNT
MLBr_01723|M.leprae_Br4923 LIQSGVARRGSVTDLSPGMVKIATRNGQSLGLDINGRVADAEGIPYDDNT
*:*:****************::***.*: ****::******* ***:*::
Mb3064c|M.bovis_AF2122/97 FDLVVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAGEPTTVGDGYARTL
Rv3038c|M.tuberculosis_H37Rv FDLVVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAGEPTTVGDGYARTL
MMAR_1663|M.marinum_M FDLVVGHAVLHHIPDVELSLREVMRVLKPGGRFVFAGEPTTVGNEYARTL
MUL_1900|M.ulcerans_Agy99 FDLVVGHAVLHHIPDVELSLREVMRVLKPGGRFVFAGEPTTVGNEYARTL
Mflv_4269|M.gilvum_PYR-GCK FDLVVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAGEPTTVGNRYAREL
Mvan_2090|M.vanbaalenii_PYR-1 FDLVVGHAVLHHIPDVELSLREVVRVLKPGGRFVFAGEPTTVGNRYAREL
MSMEG_2329|M.smegmatis_MC2_155 FDLVVGHAVLHHIPDVELSLREVVRILKPGGRFVFAGEPTTVGNRYAREL
TH_2189|M.thermoresistible__bu FDLVVGHAVLHHIPDVELSLREVVRVLRPGGRFVFAGEPTTVGNSYARAL
MAV_3903|M.avium_104 FDLVVGHAVLHHIPDVELSLREVIRVLRPGGRFVFAGEPTSAGDVYAREL
MAB_3381c|M.abscessus_ATCC_199 FDLVVGHAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTTVGNLYARAL
MLBr_01723|M.leprae_Br4923 FDLVVGHAVLHHIPDVELSLREVLRVLKPGGRFVFAGEPTDVGNRYARVF
***********************:*:*:************ .*: *** :
Mb3064c|M.bovis_AF2122/97 STLTWRVVTNATKLPGLRGWRRPQGELDESSRAAALEALVDLHTFTPQDL
Rv3038c|M.tuberculosis_H37Rv STLTWRVVTNATKLPGLRGWRRPQGELDESSRAAALEALVDLHTFTPQDL
MMAR_1663|M.marinum_M STLTWRIATNVTKLPGLGGWRRPQEELDESSRAAALEAIVDLHTFAPEDL
MUL_1900|M.ulcerans_Agy99 STLTWRIATNVTKLPGLGGWRRPQEELDESSRAAALEAIVDLHTFAPEDL
Mflv_4269|M.gilvum_PYR-GCK STLTWHAATNIMKLPMLSDWRRPQAELDESSRAAALEAVVDLHTFDPADL
Mvan_2090|M.vanbaalenii_PYR-1 STLTWHAATTLTKLPWLDGWRRPQAELDESSRAAALEAVVDLHTFDPADL
MSMEG_2329|M.smegmatis_MC2_155 STLTWHATTNLTKLPFLSGWRRPQEELDESSRAAALEAVVDLHTFDPADL
TH_2189|M.thermoresistible__bu STLTWHVATTVTRLPGLQGWRRPQAELDESSRAAALEAVVDLHTFEPSDL
MAV_3903|M.avium_104 STLTWRIATNVTKLPGLGSWRRPQAELDESSRAAALEAIVDLHTFTPGDL
MAB_3381c|M.abscessus_ATCC_199 ADLTWKATVQAMKLPGLESWRRPQEELDENSRAAALEWVVDLHTFDPADL
MLBr_01723|M.leprae_Br4923 ADLTWKVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVL
: ***: . :** * ***.* *:**.******* :****** * *
Mb3064c|M.bovis_AF2122/97 QRIAHNAGAVEVQTATEEFTAAMLGWPLRTFECTVPPGRLGWGWARFAFT
Rv3038c|M.tuberculosis_H37Rv QRIAHNAGAVEVQTATEEFTAAMLGWPLRTFECTVPPGRLGWGWARFAFT
MMAR_1663|M.marinum_M ERMAKNAGAIRVETASEEFSAAMFGWPVRTFESSVPPGRLGWNWARFAFN
MUL_1900|M.ulcerans_Agy99 ERMAKNAGAIRVETASEEFSAAMFGWPVRTFESSVPPGRLGWNWARFAFN
Mflv_4269|M.gilvum_PYR-GCK ERMAANAGAVEVRTASEEFTAAMLGWPVRTFEAAVPPGRLGWGWAKFAFG
Mvan_2090|M.vanbaalenii_PYR-1 ERMAVNAGAVEVRTASEEFTAAMLGWPIRTFEAAVPPGRLGWGWAKFAFG
MSMEG_2329|M.smegmatis_MC2_155 ERMATNAGAVEVRTASEEFTAAMLGWPVRTFEAAVPPGRLGWGWAKFAFN
TH_2189|M.thermoresistible__bu ERMARNAGAVEVATASEEFTAAMLGWPVRTFEAAVPPGKLGWNWAKFAFG
MAV_3903|M.avium_104 ERMAANAGATEVRTVSEEFTAAMFGWPVRTFEASVPPGRLGWGWAKFAFN
MAB_3381c|M.abscessus_ATCC_199 EKMAANAGAVAVRTASEEFTAAMLGWPVRTFESTVPPGKLGWGWAKFAFN
MLBr_01723|M.leprae_Br4923 ETMATNAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFT
: :* **** * *.:***:***:***:****.::**.:***.**:***
Mb3064c|M.bovis_AF2122/97 SWKTLGWVDANVWRHVVPKGWFYNVMITGVKPS
Rv3038c|M.tuberculosis_H37Rv SWKTLGWVDANVWRHVVPKGWFYNVMITGVKPS
MMAR_1663|M.marinum_M GWKALSWMDANVWRHVAPKGWFYNVMVTGVKPS
MUL_1900|M.ulcerans_Agy99 GWKALGWMDANVWRHVAPKGWFYNVMVTGVKPS
Mflv_4269|M.gilvum_PYR-GCK SWTTLSWVDANVWRRIVPKGWFYNVMVTGVKPS
Mvan_2090|M.vanbaalenii_PYR-1 SWTTLSWVDANIWRRVVPKGWFYNVMVTGVKPS
MSMEG_2329|M.smegmatis_MC2_155 SWTTLSWVDANVWRKVVPKGWFYNVMVTGVKPS
TH_2189|M.thermoresistible__bu SWTALSWVDANVWRRVMPKGWFYNVMVTGVKPS
MAV_3903|M.avium_104 GWKTLSWVDANIWRRVVPKGWFYNVMVTGVKPT
MAB_3381c|M.abscessus_ATCC_199 GWKALSWVDENVWRHVVPKGWYYNVMITGIKPS
MLBr_01723|M.leprae_Br4923 SWKTLSWVDANVWRRVIPKSWFYNIIITGVKPS
.*.:*.*:* *:**:: **.*:**:::**:**: