For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSRTFGAVDALAPCDLAVKRGEFISIIGPSGCGKSTLLRLASKLDEPTAGAVEFATDSVGYVFQDATLMP WRTVRRNVGLLAELQGRGRAEIRSEVESALATVGLTDFAEHLPRQLSGGMKMRASLARSLVLKPEVFLFD EPFGALDEITRGRLNLVLQDLFLQHSFAGLFVTHSVEEAVFLSNRVVVMSARPGRIVDEFTVPFEFPRAL ELRYQPEFAELAGRVAKSLEEAS
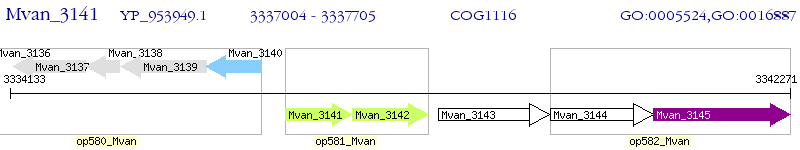
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3141 | - | - | 100% (233) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_0123 | - | 6e-40 | 38.06% (247) | ABC transporter-related protein |
| M. vanbaalenii PYR-1 | Mvan_3387 | - | 2e-34 | 40.30% (201) | spermidine/putrescine ABC transporter ATPase subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 3e-31 | 37.56% (205) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_0722 | - | 5e-39 | 41.00% (239) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 3e-31 | 37.56% (205) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01424 | - | 3e-27 | 31.25% (224) | putative binding-protein-dependent transport protein |
| M. abscessus ATCC 19977 | MAB_2178 | - | 2e-39 | 38.24% (238) | ABC transporter ATP-binding protein |
| M. marinum M | MMAR_3715 | cysA1 | 1e-30 | 38.24% (204) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. avium 104 | MAV_2434 | - | 2e-37 | 40.61% (229) | putative aliphatic sulfonates transport ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_2502 | - | 6e-51 | 44.87% (234) | transporter ATPase |
| M. thermoresistible (build 8) | TH_3596 | - | 1e-35 | 44.29% (219) | PUTATIVE ABC transporter related |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 1e-30 | 38.24% (204) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2419c|M.bovis_AF2122/97 --------MT------YAIVVADATKRYGD----FVALDHVDFVVPTGSL
Rv2397c|M.tuberculosis_H37Rv --------MT------YAIVVADATKRYGD----FVALDHVDFVVPTGSL
MMAR_3715|M.marinum_M --MKKETSLTGPGTGGHAIVVRDAFKQYGD----FVALDHVDFVVPAGSL
MUL_3658|M.ulcerans_Agy99 --------MTGPGTGGHAIVVRDAFKQYGD----FVALDHVDFVVPAGSL
Mvan_3141|M.vanbaalenii_PYR-1 MS-----------------------RTFGA----VDALAPCDLAVKRGEF
MSMEG_2502|M.smegmatis_MC2_155 MTDQRSVRPIV--------EVSGCVQRFGG----LHALGPIDLTIRSGEF
Mflv_0722|M.gilvum_PYR-GCK MSSQSDTEPVAPQRRSGAIQIRSVSHRYGQGRDEVTALGPVDLTVEPGSF
MAB_2178|M.abscessus_ATCC_1997 MPLVDEAETAVQERDTGSIRISGVTHSYGSGAGKTTALGPVDLTVEPGTF
MAV_2434|M.avium_104 MTLTAESGPLAGSRTDVAGELRHVDKWYGNR----HVLQDVSLQIPSGQI
TH_3596|M.thermoresistible__bu --MAAGPGRLTGNRAEPVAVVRGLHRSFGAG----AVLDGIDLTIRRGEF
MLBr_01424|M.leprae_Br4923 ---------------MASVTFEQATRQFPDTDR--PALDRLDLDVDDGEF
: : .* .: : * :
Mb2419c|M.bovis_AF2122/97 TALLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRRGIGFVF
Rv2397c|M.tuberculosis_H37Rv TALLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVPPQRRGIGFVF
MMAR_3715|M.marinum_M TALLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRRGIGFVF
MUL_3658|M.ulcerans_Agy99 TALLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVPPQRRGIGFVF
Mvan_3141|M.vanbaalenii_PYR-1 ISIIGPSGCGKSTLLRLASKLDEPTAGAVEFA----------TDSVGYVF
MSMEG_2502|M.smegmatis_MC2_155 VSIVGPSGCGKSTLLELVAGLQVPADGRVSVAGE---PLRGPRDRTAIAF
Mflv_0722|M.gilvum_PYR-GCK LVLVGASGCGKSTLLRLLAGFESPTDGSVDVEG----VPPTPGVTAGVVF
MAB_2178|M.abscessus_ATCC_1997 LVLVGASGCGKSTLLRLLAGFESPSEGAVRVAG----GVPTPGITSGVVF
MAV_2434|M.avium_104 VALIGRSGSGKSTVLRVLAGLSHDHTGRR-----------LVAGAPALAF
TH_3596|M.thermoresistible__bu VALLGRSGSGKSTLLRALAGLDHGGAGGGGGDV-------YVSRDVAVVF
MLBr_01424|M.leprae_Br4923 VVVVGPSGCGKTTSLRMVAGLETLDSGCIRIGDRDVTHVDPKDRDVAMVF
::* **.**:* *. : :. * . .*
Mb2419c|M.bovis_AF2122/97 QHYAAFKHLTVRDNVAFGLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYP
Rv2397c|M.tuberculosis_H37Rv QHYAAFKHLTVRDNVAFGLKIRKRPKAEIKAKVDNLLQVVGLSGFQSRYP
MMAR_3715|M.marinum_M QHYAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYP
MUL_3658|M.ulcerans_Agy99 QHYAAFKHLTVRDNVAYGLKIRKRPKAEIRDKVDNLLEVVGLSGFHGRYP
Mvan_3141|M.vanbaalenii_PYR-1 QDATLMPWRTVRRNVGLLAELQGRGRAEIRSEVESALATVGLTDFAEHLP
MSMEG_2502|M.smegmatis_MC2_155 QESATLPWRTVRDNVAFALEVRAVPKAERRRRADELIRTVGLAGFEDNYP
Mflv_0722|M.gilvum_PYR-GCK QQPRLFPWRTVGGNVELALKYAKVPRDKRSERRDELLERVGLGGTAGRRI
MAB_2178|M.abscessus_ATCC_1997 QQPRLFPWRTVGGNIELALKYAKVPRERWAQRRDQLLARVGLEGTAGRRI
MAV_2434|M.avium_104 QEPRLFPWRDVRTNVGYGLTRTRLPRAQVRRRAERALADVGLADHARAWP
TH_3596|M.thermoresistible__bu QDSRLLPWARVLDNVTLGLSGR-----DAERRGAAVLADVGLAGRERAWP
MLBr_01424|M.leprae_Br4923 QNYALYPHMTVAQNLGFALKISKISKAEIHDRILVVAKLLDLQPYLDRKP
*. * *: . :.*
Mb2419c|M.bovis_AF2122/97 NQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHD
Rv2397c|M.tuberculosis_H37Rv NQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREELRAWLRRLHD
MMAR_3715|M.marinum_M NQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHD
MUL_3658|M.ulcerans_Agy99 DQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAKVREDLRAWLRRLHD
Mvan_3141|M.vanbaalenii_PYR-1 RQLSGGMKMRASLARSLVLKPEVFLFDEPFGALDEITRGRLNLVLQDLFL
MSMEG_2502|M.smegmatis_MC2_155 SQLSGGMRQRVAIARCLSMDPDLILADEPFGALDEQTRLLMSFELMRVVE
Mflv_0722|M.gilvum_PYR-GCK WEISGGQQQRVAIARALAAETPLFLLDEPFAALDALTRERLQEDVRQVSA
MAB_2178|M.abscessus_ATCC_1997 WEISGGQQQRVAIARALAAETPLFLLDEPFAALDALTRERLQEDVRQVSA
MAV_2434|M.avium_104 LTLSGGQAQRVSLARALVAEPRLLLLDEPFGALDALTRLSMHTLLLDLWR
TH_3596|M.thermoresistible__bu YELSGGEQQRVALARSLVRDPALLLADEPFGALDALTRIRMHRLLQELCA
MLBr_01424|M.leprae_Br4923 KDLSGGQRQRVAMGRAIVRRPQVFLMDEPLSNLDAKLRAQTRNQIAELQC
:*** * ::.*.: . ::* ***:. ** * : :
Mb2419c|M.bovis_AF2122/97 EVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVM
Rv2397c|M.tuberculosis_H37Rv EVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGSPTDVYDAPANAFVM
MMAR_3715|M.marinum_M EVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPANAFVM
MUL_3658|M.ulcerans_Agy99 EVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGSPTEVYDAPTNAFVM
Mvan_3141|M.vanbaalenii_PYR-1 QHSFAGLFVTHSVEEAVFLSNRVVVMSAR-----------PGRIVDEFTV
MSMEG_2502|M.smegmatis_MC2_155 ELSCGVLFITHSIQEAVLLSDRVLVMSAR-----------PGTILDEIHV
Mflv_0722|M.gilvum_PYR-GCK ESGRTTVFVTHSADEAAFLGSRIVVLTRR-----------PGQIALDIPV
MAB_2178|M.abscessus_ATCC_1997 ESGRTTVFVTHSADEAAFLGSRIVVLTRR-----------PGQVALDIPV
MAV_2434|M.avium_104 RHGFGVLLVTHDVDEAVALADRVLVLED-------------GRVVHELAI
TH_3596|M.thermoresistible__bu KYRPGVLLVTHDVDEAVTLADRVLVLAD-------------GRFVTDREL
MLBr_01424|M.leprae_Br4923 QLGTTTVYVTHDQVEAMTMGDRVAVLCYGVLQQFATPRELYHKPENVFVA
. : :**. ** :..*: *:
Mb2419c|M.bovis_AF2122/97 SFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVV
Rv2397c|M.tuberculosis_H37Rv SFLGAVSTLNGSLVRPHDIRVGRTPNMAVAAADGTAGSTGVLRAVVDRVV
MMAR_3715|M.marinum_M SFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIV
MUL_3658|M.ulcerans_Agy99 SFLGAVSALNGALVRPHDIRVGRTPEMAVAAADGTGESTGVTRAVVDRIV
Mvan_3141|M.vanbaalenii_PYR-1 PFEFP--RALELRYQPEFAELAGRVAKSLEEAS-----------------
MSMEG_2502|M.smegmatis_MC2_155 DLPRP--RDEHTLASPRMAALTERIWAQLKDEAGKAMEATLR--------
Mflv_0722|M.gilvum_PYR-GCK DLPRTGVDPEELRRSPEYGELRAEVGAAVRRAAA----------------
MAB_2178|M.abscessus_ATCC_1997 DLPRTGIDPDDLRRSPEYAQLRTEVGRAVKAAAA----------------
MAV_2434|M.avium_104 DPP-RRTPGEPG---AHTERYRAELLDRLGVRQ-----------------
TH_3596|M.thermoresistible__bu TFRGRRTLRNP-----EFVTHREALLAALGVGDD----------------
MLBr_01424|M.leprae_Br4923 GFIGSPGMNLVTLSIVDSSVLLGDWPIRIPREIASAASEVIIGVRPEHFE
:
Mb2419c|M.bovis_AF2122/97 VLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIA
Rv2397c|M.tuberculosis_H37Rv VLGFEVRVELTSAATGGAFTAQITRGDAEALALREGDTVYVRATRVPPIA
MMAR_3715|M.marinum_M VLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIA
MUL_3658|M.ulcerans_Agy99 VLGFEVRVELTSAATGGAFTAQITRGDAEALALQEGDTVYVRATRVPPIA
Mvan_3141|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2502|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0722|M.gilvum_PYR-GCK --------------------------------------------------
MAB_2178|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_2434|M.avium_104 --------------------------------------------------
TH_3596|M.thermoresistible__bu --------------------------------------------------
MLBr_01424|M.leprae_Br4923 LGSLGVEVEIDMVEELGADAYLYGRIAGASKVTDQLVVARVDGRNPPAKG
Mb2419c|M.bovis_AF2122/97 GGVSGVDDAGVERVKVTST------
Rv2397c|M.tuberculosis_H37Rv GGVSGVDDAGVERVKVTST------
MMAR_3715|M.marinum_M GAAPAVPDDGAGSTPSAAQIGVGGQ
MUL_3658|M.ulcerans_Agy99 GAAPAVPDDGAGSTPRAAQIGVGGQ
Mvan_3141|M.vanbaalenii_PYR-1 -------------------------
MSMEG_2502|M.smegmatis_MC2_155 -------------------------
Mflv_0722|M.gilvum_PYR-GCK -------------------------
MAB_2178|M.abscessus_ATCC_1997 -------------------------
MAV_2434|M.avium_104 -------------------------
TH_3596|M.thermoresistible__bu -------------------------
MLBr_01424|M.leprae_Br4923 SRVRLYTEPGNVHFFGVDGHRIS--