For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KYLDEFSNPELAARLIEQITSAATRRWSIMEVCGGQTHSIIRHGIDQLLPDTVEMIHGPGCPVCVTPLEI IDKALDIASRPDVVFCSFGDMLRVPGSEKDLFRVKSEGGDVRVVYSPLDALTIAREHPDKQVVFFGIGFE TTAPANAMTVYQARRLGIDNFSLLVSHVLVPPAISAIMESPTCRVQAFLAAGHVCAVMGTDEYPPLCDKY GIPIVVTGFEPLDILEGIRRTVVQLESGRHELENAYPRAVREAGNPAAKAMLLDVFELTDRAWRGIGVIP GSGWRLSHAYRDYDAEQRFAVSDIHTEESAVCRSGEVLQGLIKPHECAAFGTVCTPRNPLGATMVSSEGA CAAYYLYRRLEVTNA*
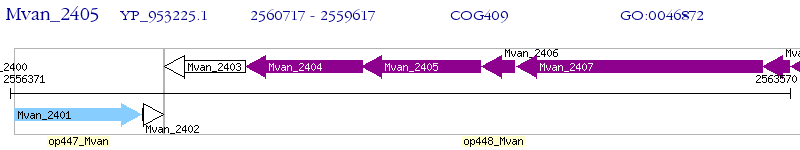
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2405 | - | - | 100% (366) | hydrogenase expression/formation protein HypD |
| M. vanbaalenii PYR-1 | Mvan_2361 | - | e-150 | 69.61% (362) | hydrogenase expression/formation protein HypD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3981 | - | 0.0 | 89.07% (366) | hydrogenase expression/formation protein HypD |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1857 | hypD | 1e-106 | 51.36% (368) | hydrogenase-forming protein, HypD |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2275 | hypD | 1e-151 | 69.89% (362) | hydrogenase expression/formation protein HypD |
| M. thermoresistible (build 8) | TH_0227 | hypD | 1e-105 | 51.23% (367) | hydrogenase expression/formation protein HypD |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2405|M.vanbaalenii_PYR-1 MKYLDEFSNPELAARLIEQITSAATR------RWSIMEVCGGQTHSIIRH
Mflv_3981|M.gilvum_PYR-GCK MKYLDEFSNPELAHRLIEQIEAVATR------RWSIMEVCGGQTHSIIRH
MSMEG_2275|M.smegmatis_MC2_155 MKYLDEFRDPVAARTLVDRINRAATR------TWTMMEVCGGQTHSIIRN
MMAR_1857|M.marinum_M MKFVDEFRDPAAARKLLTAIEHLAHQPGAGPEHFKFMEVCGGHTHTIYRH
TH_0227|M.thermoresistible__bu MRFVDEFRDPAAARKLLAAIEVLAAG-GEDGAPFKFMEVCGGHTHTIYRH
*:::*** :* * *: * * :.:******:**:* *:
Mvan_2405|M.vanbaalenii_PYR-1 GIDQLLPDTVEMIHGPGCPVCVTPLEIIDKALDIASRPDVVFCSFGDMLR
Mflv_3981|M.gilvum_PYR-GCK GIDQLLPDTVEMIHGPGCPVCVTPLEIIDKALDIASRPEVIFCSFGDMLR
MSMEG_2275|M.smegmatis_MC2_155 GIDQLLEGSVEFVHGPGCPVCVTPLEVIDRALEIAARDDVIFCSFGDMLR
MMAR_1857|M.marinum_M GIEHLLPDNVELVHGPGCPVCVIPMGRIDDAMWLAGQPDVIFTCFGDMMR
TH_0227|M.thermoresistible__bu GIEHLLPPTVELVHGPGCPVCVIPMGRIDDAMWLAAQPDVIFTSFGDMMR
**::** .**::********* *: ** *: :*.: :*:* .****:*
Mvan_2405|M.vanbaalenii_PYR-1 VPGSEKDLFRVKSEGGDVRVVYSPLDALTIAREHPDKQVVFFGIGFETTA
Mflv_3981|M.gilvum_PYR-GCK VPGSEKDLFRVKSEGADVRVVYSPLDALTIARDNPDRQVVFFGIGFETTA
MSMEG_2275|M.smegmatis_MC2_155 VPGSHQDLFSVKARGGDVRIVYSPLDATRVAADNPDKQVVFFGVGFETTA
MMAR_1857|M.marinum_M VPGSDGSLLDAKARGADVRFVYSPLDALKVAIDNPDKQVVFFAIGFETTA
TH_0227|M.thermoresistible__bu VPGSNGSLLDAKARGADVRFVYSPLDALKIAVDNPDKRVVYFAIGFETTA
****. .*: .*:.*.***.******* :* ::**::**:*.:******
Mvan_2405|M.vanbaalenii_PYR-1 PANAMTVYQARRLGIDNFSLLVSHVLVPPAISAIMESPTCRVQAFLAAGH
Mflv_3981|M.gilvum_PYR-GCK PANAMTVYQAKKLGIENFSLLVSHVLVPPAIAAIMESPTCRVQAFLAAGH
MSMEG_2275|M.smegmatis_MC2_155 PANAMAVVHAKRLGLKNFSMLVSHVLVPPAMTAILSSPSNRVQGFLAAGH
MMAR_1857|M.marinum_M PSTAVTLVRARDLELPNFSVFCNHVTIVPPIKAILESPDLRLSGFIGPGH
TH_0227|M.thermoresistible__bu PSTAVTLLRATELGLRNFSVFCNHVTIVPPIKAILESPDLRLSGFLGPGH
*:.*::: :* * : ***:: .** : *.: **:.** *:..*:..**
Mvan_2405|M.vanbaalenii_PYR-1 VCAVMGTDEYPPLCDKYGIPIVVTGFEPLDILEGIRRTVVQLESGRHELE
Mflv_3981|M.gilvum_PYR-GCK VCTVMGTDEYPPLCDKYGTPIVVTGFEPLDILEGIRRTVIQLEAGRHELE
MSMEG_2275|M.smegmatis_MC2_155 VCSVMGTSEYGPLVEKFDVPIVVTGFEPLDLLEGVRQLVDLLEAGTPELR
MMAR_1857|M.marinum_M VSTVVGNRPYRFVPEVYHKPLVVAGFEPLDILASVTMLLRQIREGRCEVE
TH_0227|M.thermoresistible__bu VSTVVGIRPYRFVPEVYRKPLVVAGFEPLDILAAVAMLLRQIREGRCEVE
*.:*:* * : : : *:**:******:* .: : :. * *:.
Mvan_2405|M.vanbaalenii_PYR-1 NAYPRAVREAGNPAAKAMLLDVFELTDR-AWRGIGVIPGSGWRLSHAYRD
Mflv_3981|M.gilvum_PYR-GCK NAYPRAVQDTGNPAAKAMLLDVFDVTDR-TWRGIGMIASSGWRLSPAYRH
MSMEG_2275|M.smegmatis_MC2_155 NAYSRAVTAEGNVFAQQTLSEVFTVTDR-QWRGIGMIPRSGWTLSPQYAE
MMAR_1857|M.marinum_M NQYKRVVPEEGNPAALALMGKVFALRPHFEWRGLGFISQSALRLSDDFAE
TH_0227|M.thermoresistible__bu NQYSRIVAPQGNPAALGMLARVFELRPHFEWRGLGFISQSALKLRDEFAD
* * * * ** * : ** : : ***:*.*. *. * : .
Mvan_2405|M.vanbaalenii_PYR-1 YDAEQRFAVSDIHTEESAVCRSGEVLQGLIKPHECAAFGTVCTPRNPLGA
Mflv_3981|M.gilvum_PYR-GCK FDAEHRFAATDLHNEESAICRSGEVLQGLIKPHECTAFGTSCTPRNPLGA
MSMEG_2275|M.smegmatis_MC2_155 FDAELRFGVGHLQVAESAECRSGEVLQGLLKPNECPAFGKTCTPRNPLGA
MMAR_1857|M.marinum_M FDAELRFAMPGVRVADPKACQCGEVLKGVLKPWECKVFGTACTPETPIGT
TH_0227|M.thermoresistible__bu YDAERLFDMPGVRVADPKACQCGEVLKGVLRPWECKVFGTACTPETPIGT
:*** * :: :. *:.****:*:::* ** .**. ***..*:*:
Mvan_2405|M.vanbaalenii_PYR-1 TMVSSEGACAAYYLYRRLEVTNA---------
Mflv_3981|M.gilvum_PYR-GCK TMVSSEGACAAYYLYRRLEVTHA---------
MSMEG_2275|M.smegmatis_MC2_155 TMVSSEGACAAYYQFRRLEAAAHV--------
MMAR_1857|M.marinum_M CMVSPEGACAAYYNFGRLHRDAVKLVGQSGRA
TH_0227|M.thermoresistible__bu CMVSSEGACAAYYNFGRLHRDAATLVGQAGRG
***.******** : **.