For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAEPVAPASSRRIAALAFPALGVLAAEPIYLLFDLAVIGRLGALNLAGLAIGALIMGVLSSQLTFLSYGT TARAARFYGAGNRTAAVEEGVQATWLALGIGTTIVVAVQLTAVPLVSVLAGGADHGRIAETALPWVRIAS LAVPAILIAAAGNGWMRGVQDTMRPLRYVIFGFAVSAVLCPLLVYGWLGAPELGLPGSAVANVVGQYLAA ALFCRALVVEKVPLRLRPPVLRAQVVMGRDLVLRTMAFQACFISAGAVAARFGAAAVAAHQVVLQLWNFL ALVLDSLAIAAQSLVGAALGAGQLTHAKAVAWRVTIFSTVAGVVLATVFALGSSVFPAVFTDDRSVLDQI GVPWWFLVAQLPVAGIVFAIDGVLLGAGDATFMRNATLISALVGFLPLIWLSLAFGWGLLGIWAGLSAFM VLRLVFVGWRALSGRWLVPGTR
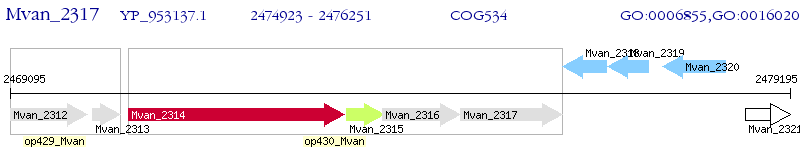
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2317 | - | - | 100% (442) | MATE efflux family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2861c | dinF | 1e-177 | 71.82% (433) | DNA-damage-inducible protein F |
| M. gilvum PYR-GCK | Mflv_4036 | - | 0.0 | 89.59% (442) | MATE efflux family protein |
| M. tuberculosis H37Rv | Rv2836c | dinF | 1e-179 | 72.06% (433) | DNA-damage-inducible protein F |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3128c | - | 1e-168 | 67.36% (435) | DNA-damage-inducible protein F |
| M. marinum M | MMAR_1897 | dinF | 1e-178 | 71.95% (435) | DNA-damage-inducible protein F DinF |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2631 | - | 0.0 | 80.88% (434) | MATE efflux family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2125 | dinF | 1e-177 | 71.72% (435) | DNA-damage-inducible protein F DinF |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2317|M.vanbaalenii_PYR-1 MAEP----------------VAPASSRRIAALAFPALGVLAAEPIYLLFD
Mflv_4036|M.gilvum_PYR-GCK MADP----------------VAPATGRRIAGLAFPALGVLAAEPVYLLFD
MSMEG_2631|M.smegmatis_MC2_155 MADAAGDLPGGPDDDAQGAALTQATGRRIAKLAFPALGVLAAEPIYLLFD
MMAR_1897|M.marinum_M MGPDAGRP--------------DAGGRKIAALALPALGVLAAEPLYLLFD
MUL_2125|M.ulcerans_Agy99 MGPDAGRP--------------DAGGRKIAALALPALGVLAAEPLYLLFD
Mb2861c|M.bovis_AF2122/97 MSQVGHR----------------AGGRQIAQLALPALGVLAAEPLYLLFD
Rv2836c|M.tuberculosis_H37Rv MSQVGHR----------------AGGRQIAQLALPALGVLAAEPLYLLFD
MAB_3128c|M.abscessus_ATCC_199 MTDATGIAG------------MPGLARRIISLALPALGVLAAEPLYLLFD
* . .*:* **:**********:*****
Mvan_2317|M.vanbaalenii_PYR-1 LAVIGRLGALNLAGLAIGALIMGVLSSQLTFLSYGTTARAARFYGAGNRT
Mflv_4036|M.gilvum_PYR-GCK LAIVGRLGALSLAGLAIGALIMGVLSSQLTFLSYGTTARAARFYGAGDRN
MSMEG_2631|M.smegmatis_MC2_155 LAIVGRLGAVSLAGLAIGGLVLGLVNSQGTFLSYGTTARSARFYGAGDRT
MMAR_1897|M.marinum_M TAVIGRLGALALAGLAIGSLVLGLVGSQATFLSYGTTARSARHYGAGDRV
MUL_2125|M.ulcerans_Agy99 TAVIGRLGALALAGLAIGSLVLGLVGSQATFLSYGTTARSARHYGAGDRV
Mb2861c|M.bovis_AF2122/97 IAVVGRLGAISLAGLAIGSLVLGLVGSQATFLSYGTTARAARRYGAGNRV
Rv2836c|M.tuberculosis_H37Rv IAVVGRLGAISLAGLAIGSLVLGLVGSQATFLSYGTTARAARRYGAGNRV
MAB_3128c|M.abscessus_ATCC_199 IAMVGRLGAVPLAGLAVGGLVLSLVGTQLTFLSYGTTARAARRFGSGDRP
*::*****: *****:*.*::.::.:* **********:** :*:*:*
Mvan_2317|M.vanbaalenii_PYR-1 AAVEEGVQATWLALGIGTTIVVAVQLTAVPLVSVLAGGAD-HGRIAETAL
Mflv_4036|M.gilvum_PYR-GCK AAVEEGVQATWLAVGIGTVIVAAVQVTAVPLVSALAAG----GEIAETAL
MSMEG_2631|M.smegmatis_MC2_155 SAVAEGVQATWLALGLGLLIIAVVEAVAVPMLSALAAG----GEIAHAAL
MMAR_1897|M.marinum_M AAVAEGVQATWLAVALGLLTILVVQIAAVPLVSVIAGARAGSGDIAGAAL
MUL_2125|M.ulcerans_Agy99 AAVAEGVQATWLAVALGLLTILVVQIAAVPLVSVIAGARAGSGDIAGAAL
Mb2861c|M.bovis_AF2122/97 AAVTEGVQATWLALGLGALVVVVVEATATPLVSAIA---SGDG-ITAAAL
Rv2836c|M.tuberculosis_H37Rv AAVTEGVQATWLALGLGALVVVVVEATATPLVSAIA---SGDG-ITAAAL
MAB_3128c|M.abscessus_ATCC_199 GAVHEGVQATWLALLIGAVVVLVVHAVAGPVVRAIAAA----PDVAAQGL
.** *********: :* : .*. .* *:: .:* :: .*
Mvan_2317|M.vanbaalenii_PYR-1 PWVRIASLAVPAILIAAAGNGWMRGVQDTMRPLRYVIFGFAVSAVLCPLL
Mflv_4036|M.gilvum_PYR-GCK PWVRIASLAVPAILIAAAGNGWMRGVQDTMRPLRYVVVGFGLSAVLCPLL
MSMEG_2631|M.smegmatis_MC2_155 PWLRVAILAAPAILVSMAGNGWMRGVQDTVRPLRYVVLGFAVSAVLCPLL
MMAR_1897|M.marinum_M PWLRIAILGTPAILISLAGNGWMRGVQDTVRPLRYVVAGFGLSALLCPLL
MUL_2125|M.ulcerans_Agy99 PWLWIAILGTPAILISLAGNGWMRGVQDTVRPLRYVVAGFGLSALLCPLL
Mb2861c|M.bovis_AF2122/97 PWLRIAILGTPAILVSLAGNGWLRGVQDTVRPLRYVVAGFGSSALLCPLL
Rv2836c|M.tuberculosis_H37Rv PWLRIAILGTPAILVSLAGNGWLRGVQDTVRPLRYVVAGFGSSALLCPLL
MAB_3128c|M.abscessus_ATCC_199 GWLRIAIFAAPAILVSLAGNGWMRGVQNTVRPLRYVIAGFAVSAVLCPVL
*: :* :..****:: *****:****:*:******: **. **:***:*
Mvan_2317|M.vanbaalenii_PYR-1 VYGWLGAPELGLPGSAVANVVGQYLAAALFCRALVVEKVPLRLRPPVLRA
Mflv_4036|M.gilvum_PYR-GCK VFGWLGCPELGLPGSAVANVAGQYLAAALFCRALFVEKVPLRVRPAVLRA
MSMEG_2631|M.smegmatis_MC2_155 VYGWLGFPRLELAGSAVANVVGQWLAAGLFMRALLVEKVPLRLRPDVLRV
MMAR_1897|M.marinum_M VYGWLGLPRMELAGSAVANLVGQWLAALLFGGALLAERVSLRPDRHILRE
MUL_2125|M.ulcerans_Agy99 VYGWLGLPRMELAGSAVANLVGQWLAALLFGGALLAERVSLRPDRHILRE
Mb2861c|M.bovis_AF2122/97 VYGWLGLPRWGLTGSAVANLVGQWLAALLFAGALLAERVSLRPDRAVLGA
Rv2836c|M.tuberculosis_H37Rv VYGWLGLPRWGLTGSAVANLVGQWLAALLFAGALLAERVSLRPDRAVLGA
MAB_3128c|M.abscessus_ATCC_199 IYGLAGVPRMGLAGSAVANLVGQWLAAILFLRALHAEHAPLRTDRAVLRA
::* * *. *.******:.**:*** ** ** .*:..** :*
Mvan_2317|M.vanbaalenii_PYR-1 QVVMGRDLVLRTMAFQACFISAGAVAARFGAAAVAAHQVVLQLWNFLALV
Mflv_4036|M.gilvum_PYR-GCK QVVMGRDLILRTMAFQACFISAGAVAARFGAASVAAHQVVLQLWSFLALV
MSMEG_2631|M.smegmatis_MC2_155 QLVMGRDLLLRSLAFQACFVSAGAVAARFGAAAVAAHQVVLQLWSFLALV
MMAR_1897|M.marinum_M QLVLARDLIVRTMAFQACFISAAAVAARFGAAALAAHQVVLQLWGFFALV
MUL_2125|M.ulcerans_Agy99 QLVLARDLIVRTMAFQACFISAAAVAARFGAAALAAHQVVLQLWGFFALV
Mb2861c|M.bovis_AF2122/97 QLMMARDLIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWGLLALV
Rv2836c|M.tuberculosis_H37Rv QLMMARDLIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWGLLALV
MAB_3128c|M.abscessus_ATCC_199 QLVLARDLLVRSLAFQACFISAAAVAARFGAAALAAHQVVLQMWSFLALV
*:::.***::*::***.*::**.*********::********:*.::***
Mvan_2317|M.vanbaalenii_PYR-1 LDSLAIAAQSLVGAALGAGQLTHAKAVAWRVTIFSTVAGVVLATVFALGS
Mflv_4036|M.gilvum_PYR-GCK LDSLAIAAQSLVGAALGAGQLTHAKAVAWRVTIFSAVAGVVLAVVFALGS
MSMEG_2631|M.smegmatis_MC2_155 LDSLAIAAQSLVGAALGAGQLAHAKSVAWRVTLFSTLAGVVLAVVFAVGA
MMAR_1897|M.marinum_M LDSLAIAAQSLVGAALGAGDAAHAKWVAWRVTVFSLLAAGMLAAALALGA
MUL_2125|M.ulcerans_Agy99 LDSLAIAAQSLVGAALGAGDAAHAKWVAWRVTVFSLLAAGMLAAALALGA
Mb2861c|M.bovis_AF2122/97 LDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGS
Rv2836c|M.tuberculosis_H37Rv LDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGS
MAB_3128c|M.abscessus_ATCC_199 LDSLAIAAQTLVGAALGAGRVPEAKSVAWRVSIFSLGFAVLLAGILALGA
*********:********* .** *****: ** . :** :.:*:
Mvan_2317|M.vanbaalenii_PYR-1 SVFPAVFTDDRSVLDQIGVPWWFLVAQLPVAGIVFAIDGVLLGAGDATFM
Mflv_4036|M.gilvum_PYR-GCK QLIPSAFTDDQSVLDRIGVPWWFLVAQLPVAGIVFAIDGVLLGAGDATFM
MSMEG_2631|M.smegmatis_MC2_155 SVFPPVFTDDESVLATIGVPWWFLVAQLPVAGIVFALDGVLLGAGDAKFM
MMAR_1897|M.marinum_M PVLPALFTDDAAVLAAVTVPWWFLVAQLPFAGIVFALDGVLLGAGDAAFM
MUL_2125|M.ulcerans_Agy99 PVLPALFTDDAAVLAAITVPWWFLVAQLPFAGVVFALDGVLLGAGDAAFM
Mb2861c|M.bovis_AF2122/97 SVLPGLFTDDRSVLAAIGVLWWFMVVQLPFAGIVFAVDGVLLGAGDAAFM
Rv2836c|M.tuberculosis_H37Rv SVLPGLFTDDRSVLAAIGVPWWFMVVQLPFAGIVFAVDGVLLGAGDAAFM
MAB_3128c|M.abscessus_ATCC_199 PVLPRLFTSDAAVLHEMRVPWWFLVCQLPISGLVFALDGVLLGAADARFM
::* **.* :** : * ***:* ***.:*:***:*******.** **
Mvan_2317|M.vanbaalenii_PYR-1 RNATLISALVGFLPLIWLSLAFGWGLLGIWAGLSAFMVLRLVFVGWRALS
Mflv_4036|M.gilvum_PYR-GCK RNATLASALAGFLPLVWLSLAFGWGLLGIWAGLSTFMVLRLVFVGWRALS
MSMEG_2631|M.smegmatis_MC2_155 RNATLASAMLGFLPLIWLSLAFGWGLFGIWSGLSTFMVLRLVFVGWRAFS
MMAR_1897|M.marinum_M RTATVVSALAGFLPLTWLSLVYGWGLAGIWSGLATFIVLRLIFVGWRAMS
MUL_2125|M.ulcerans_Agy99 RTATVVSALAGFLPLTWLSLVYGWGLAGIWSGLATFIVLRLIFVGWRAMS
Mb2861c|M.bovis_AF2122/97 RTATVASALVGFLPLVWLSLAYGWGLAGIWSGLGTFIVLRLIFVGWRAYS
Rv2836c|M.tuberculosis_H37Rv RTATVASALVGFLPLVWLSLAYGWGLAGIWSGLGTFIVLRLIFVGWRAYS
MAB_3128c|M.abscessus_ATCC_199 RNATMISALCGFLPSVWLALVFGWGLAGIWCGLTLFLVLRLVLVGWRALS
*.**: **: **** **:*.:**** ***.** *:****::***** *
Mvan_2317|M.vanbaalenii_PYR-1 GRWLVPGTR-
Mflv_4036|M.gilvum_PYR-GCK GRWLVPGTR-
MSMEG_2631|M.smegmatis_MC2_155 GRWLVPGTS-
MMAR_1897|M.marinum_M GRWALTGTA-
MUL_2125|M.ulcerans_Agy99 GRWALTGTA-
Mb2861c|M.bovis_AF2122/97 GRWAVTGAA-
Rv2836c|M.tuberculosis_H37Rv GRWAVTGAA-
MAB_3128c|M.abscessus_ATCC_199 GGWAVPGTGS
* * :.*: