For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSQVGHRAGGRQIAQLALPALGVLAAEPLYLLFDIAVVGRLGAISLAGLAIGSLVLGLVGSQATFLSYGT TARAARRYGAGNRVAAVTEGVQATWLALGLGALVVVVVEATATPLVSAIASGDGITAAALPWLRIAILGT PAILVSLAGNGWLRGVQDTVRPLRYVVAGFGSSALLCPLLVYGWLGLPRWGLTGSAVANLVGQWLAALLF AGALLAERVSLRPDRAVLGAQLMMARDLIVRTLAFQVCYVSAAAVAARFGAAALAAHQVVLQLWGLLALV LDSLAIAAQSLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPGLFTDDRSVLAAIGVL WWFMVVQLPFAGIVFAVDGVLLGAGDAAFMRTATVASALVGFLPLVWLSLAYGWGLAGIWSGLGTFIVLR LIFVGWRAYSGRWAVTGAA
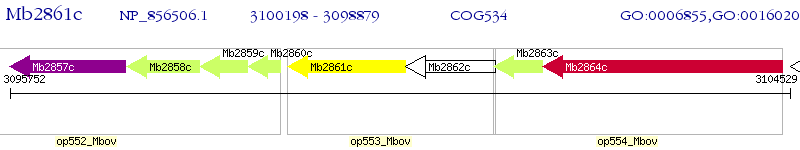
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2861c | dinF | - | 100% (439) | DNA-damage-inducible protein F |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4036 | - | 1e-179 | 72.33% (430) | MATE efflux family protein |
| M. tuberculosis H37Rv | Rv2836c | dinF | 0.0 | 99.77% (439) | DNA-damage-inducible protein F |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3128c | - | 1e-170 | 71.66% (427) | DNA-damage-inducible protein F |
| M. marinum M | MMAR_1897 | dinF | 0.0 | 83.52% (437) | DNA-damage-inducible protein F DinF |
| M. avium 104 | MAV_5113 | - | 2e-05 | 25.44% (401) | NADH dehydrogenase subunit n |
| M. smegmatis MC2 155 | MSMEG_2631 | - | 0.0 | 74.60% (433) | MATE efflux family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2125 | dinF | 0.0 | 83.30% (437) | DNA-damage-inducible protein F DinF |
| M. vanbaalenii PYR-1 | Mvan_2317 | - | 1e-177 | 71.82% (433) | MATE efflux family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2861c|M.bovis_AF2122/97 ---MSQVGHR----------------AGGRQIAQLALPALGVLAAEPLYL
Rv2836c|M.tuberculosis_H37Rv ---MSQVGHR----------------AGGRQIAQLALPALGVLAAEPLYL
MMAR_1897|M.marinum_M ---MGPDAGRP--------------DAGGRKIAALALPALGVLAAEPLYL
MUL_2125|M.ulcerans_Agy99 ---MGPDAGRP--------------DAGGRKIAALALPALGVLAAEPLYL
Mflv_4036|M.gilvum_PYR-GCK ---MADP----------------VAPATGRRIAGLAFPALGVLAAEPVYL
Mvan_2317|M.vanbaalenii_PYR-1 ---MAEP----------------VAPASSRRIAALAFPALGVLAAEPIYL
MSMEG_2631|M.smegmatis_MC2_155 ---MADAAGDLPGGPDDDAQGAALTQATGRRIAKLAFPALGVLAAEPIYL
MAB_3128c|M.abscessus_ATCC_199 ---MTDATGIAG------------MPGLARRIISLALPALGVLAAEPLYL
MAV_5113|M.avium_104 MCGIAAVFGVRQTVHVGWLLPLSGLQLDLDPVGGFFMALIGGVAVPVGFY
: : : :. :* :*. :
Mb2861c|M.bovis_AF2122/97 LFDIAVVGRLGAISLAGLAIGSLVLGLVGSQATFLSYG------TTARAA
Rv2836c|M.tuberculosis_H37Rv LFDIAVVGRLGAISLAGLAIGSLVLGLVGSQATFLSYG------TTARAA
MMAR_1897|M.marinum_M LFDTAVIGRLGALALAGLAIGSLVLGLVGSQATFLSYG------TTARSA
MUL_2125|M.ulcerans_Agy99 LFDTAVIGRLGALALAGLAIGSLVLGLVGSQATFLSYG------TTARSA
Mflv_4036|M.gilvum_PYR-GCK LFDLAIVGRLGALSLAGLAIGALIMGVLSSQLTFLSYG------TTARAA
Mvan_2317|M.vanbaalenii_PYR-1 LFDLAVIGRLGALNLAGLAIGALIMGVLSSQLTFLSYG------TTARAA
MSMEG_2631|M.smegmatis_MC2_155 LFDLAIVGRLGAVSLAGLAIGGLVLGLVNSQGTFLSYG------TTARSA
MAB_3128c|M.abscessus_ATCC_199 LFDIAMVGRLGAVPLAGLAVGGLVLSLVGTQLTFLSYG------TTARAA
MAV_5113|M.avium_104 SIGYARREHLGRVSLAMLPLFVAAMMLVPAAGSVTTFLLAWELMATASLV
:. * :** : ** *.: : :: : :. :: :** .
Mb2861c|M.bovis_AF2122/97 RRYGAGNRVAAVTEG------VQATWLALGLGALVVVVVEAT--------
Rv2836c|M.tuberculosis_H37Rv RRYGAGNRVAAVTEG------VQATWLALGLGALVVVVVEAT--------
MMAR_1897|M.marinum_M RHYGAGDRVAAVAEG------VQATWLAVALGLLTILVVQIA--------
MUL_2125|M.ulcerans_Agy99 RHYGAGDRVAAVAEG------VQATWLAVALGLLTILVVQIA--------
Mflv_4036|M.gilvum_PYR-GCK RFYGAGDRNAAVEEG------VQATWLAVGIGTVIVAAVQVT--------
Mvan_2317|M.vanbaalenii_PYR-1 RFYGAGNRTAAVEEG------VQATWLALGIGTTIVVAVQLT--------
MSMEG_2631|M.smegmatis_MC2_155 RFYGAGDRTSAVAEG------VQATWLALGLGLLIIAVVEAV--------
MAB_3128c|M.abscessus_ATCC_199 RRFGSGDRPGAVHEG------VQATWLALLIGAVVVLVVHAV--------
MAV_5113|M.avium_104 LVLAQHTRPEVRSAGLFYAVMTQLGFVAILIGLMVLSAAGGADRFADLTR
. * . * .* ::*: :* : .. .
Mb2861c|M.bovis_AF2122/97 ----ATPLVSAIA---SGDG-ITAAALPWLRIAILGTPAILVSLAGNGWL
Rv2836c|M.tuberculosis_H37Rv ----ATPLVSAIA---SGDG-ITAAALPWLRIAILGTPAILVSLAGNGWL
MMAR_1897|M.marinum_M ----AVPLVSVIAGARAGSGDIAGAALPWLRIAILGTPAILISLAGNGWM
MUL_2125|M.ulcerans_Agy99 ----AVPLVSVIAGARAGSGDIAGAALPWLWIAILGTPAILISLAGNGWM
Mflv_4036|M.gilvum_PYR-GCK ----AVPLVSALAAG----GEIAETALPWVRIASLAVPAILIAAAGNGWM
Mvan_2317|M.vanbaalenii_PYR-1 ----AVPLVSVLAGGAD-HGRIAETALPWVRIASLAVPAILIAAAGNGWM
MSMEG_2631|M.smegmatis_MC2_155 ----AVPMLSALAAG----GEIAHAALPWLRVAILAAPAILVSMAGNGWM
MAB_3128c|M.abscessus_ATCC_199 ----AGPVVRAIAAA----PDVAAQGLGWLRIAIFAAPAILVSLAGNGWM
MAV_5113|M.avium_104 VSDGARTVVFVLTMAGFGSKAGLVPLHAWLPRAHPEAPSPVSALMSAAMV
* .:: .:: *: * .*: : : . . :
Mb2861c|M.bovis_AF2122/97 RGVQDTVRPLRYVVAGFGSSALLCPLLVYG-----------WLGLPRWGL
Rv2836c|M.tuberculosis_H37Rv RGVQDTVRPLRYVVAGFGSSALLCPLLVYG-----------WLGLPRWGL
MMAR_1897|M.marinum_M RGVQDTVRPLRYVVAGFGLSALLCPLLVYG-----------WLGLPRMEL
MUL_2125|M.ulcerans_Agy99 RGVQDTVRPLRYVVAGFGLSALLCPLLVYG-----------WLGLPRMEL
Mflv_4036|M.gilvum_PYR-GCK RGVQDTMRPLRYVVVGFGLSAVLCPLLVFG-----------WLGCPELGL
Mvan_2317|M.vanbaalenii_PYR-1 RGVQDTMRPLRYVIFGFAVSAVLCPLLVYG-----------WLGAPELGL
MSMEG_2631|M.smegmatis_MC2_155 RGVQDTVRPLRYVVLGFAVSAVLCPLLVYG-----------WLGFPRLEL
MAB_3128c|M.abscessus_ATCC_199 RGVQNTVRPLRYVIAGFAVSAVLCPVLIYG-----------LAGVPRMGL
MAV_5113|M.avium_104 NLGVYGIVRFDLQLLGPGPRWWAITLLVVGGVSAVYGVLQASVATDLKRL
. : : : * . .:*: * . *
Mb2861c|M.bovis_AF2122/97 TGSAVANLVGQWLAALLFAGALLAERVSLRPDRAVLGAQLMMARDLIVRT
Rv2836c|M.tuberculosis_H37Rv TGSAVANLVGQWLAALLFAGALLAERVSLRPDRAVLGAQLMMARDLIVRT
MMAR_1897|M.marinum_M AGSAVANLVGQWLAALLFGGALLAERVSLRPDRHILREQLVLARDLIVRT
MUL_2125|M.ulcerans_Agy99 AGSAVANLVGQWLAALLFGGALLAERVSLRPDRHILREQLVLARDLIVRT
Mflv_4036|M.gilvum_PYR-GCK PGSAVANVAGQYLAAALFCRALFVEKVPLRVRPAVLRAQVVMGRDLILRT
Mvan_2317|M.vanbaalenii_PYR-1 PGSAVANVVGQYLAAALFCRALVVEKVPLRLRPPVLRAQVVMGRDLVLRT
MSMEG_2631|M.smegmatis_MC2_155 AGSAVANVVGQWLAAGLFMRALLVEKVPLRLRPDVLRVQLVMGRDLLLRS
MAB_3128c|M.abscessus_ATCC_199 AGSAVANLVGQWLAAILFLRALHAEHAPLRTDRAVLRAQLVLARDLLVRS
MAV_5113|M.avium_104 LAYSTTENMGLITLALGAATLLADSGAHSAATIAMAAALLHLIGHAAFKS
. :.:: * * * . . : : : . .::
Mb2861c|M.bovis_AF2122/97 LAFQVCYVSAAAVAAR---FGAAALAAHQVVLQLWGLLALVLDSLAIAAQ
Rv2836c|M.tuberculosis_H37Rv LAFQVCYVSAAAVAAR---FGAAALAAHQVVLQLWGLLALVLDSLAIAAQ
MMAR_1897|M.marinum_M MAFQACFISAAAVAAR---FGAAALAAHQVVLQLWGFFALVLDSLAIAAQ
MUL_2125|M.ulcerans_Agy99 MAFQACFISAAAVAAR---FGAAALAAHQVVLQLWGFFALVLDSLAIAAQ
Mflv_4036|M.gilvum_PYR-GCK MAFQACFISAGAVAAR---FGAASVAAHQVVLQLWSFLALVLDSLAIAAQ
Mvan_2317|M.vanbaalenii_PYR-1 MAFQACFISAGAVAAR---FGAAAVAAHQVVLQLWNFLALVLDSLAIAAQ
MSMEG_2631|M.smegmatis_MC2_155 LAFQACFVSAGAVAAR---FGAAAVAAHQVVLQLWSFLALVLDSLAIAAQ
MAB_3128c|M.abscessus_ATCC_199 LAFQACFISAAAVAAR---FGAAALAAHQVVLQMWSFLALVLDSLAIAAQ
MAV_5113|M.avium_104 LGFLAAGSVLAATGLRDLDLLGGLARRMPATTALFGVAALGASGLPLGAG
:.* .. .*.. * : .. .. ::.. ** ..*.:.*
Mb2861c|M.bovis_AF2122/97 SLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPGLFTD
Rv2836c|M.tuberculosis_H37Rv SLVGAALGAGDAGHAKAVAWRVTAFSLLAAGILAAALGLGSSVLPGLFTD
MMAR_1897|M.marinum_M SLVGAALGAGDAAHAKWVAWRVTVFSLLAAGMLAAALALGAPVLPALFTD
MUL_2125|M.ulcerans_Agy99 SLVGAALGAGDAAHAKWVAWRVTVFSLLAAGMLAAALALGAPVLPALFTD
Mflv_4036|M.gilvum_PYR-GCK SLVGAALGAGQLTHAKAVAWRVTIFSAVAGVVLAVVFALGSQLIPSAFTD
Mvan_2317|M.vanbaalenii_PYR-1 SLVGAALGAGQLTHAKAVAWRVTIFSTVAGVVLATVFALGSSVFPAVFTD
MSMEG_2631|M.smegmatis_MC2_155 SLVGAALGAGQLAHAKSVAWRVTLFSTLAGVVLAVVFAVGASVFPPVFTD
MAB_3128c|M.abscessus_ATCC_199 TLVGAALGAGRVPEAKSVAWRVSIFSLGFAVLLAGILALGAPVLPRLFTS
MAV_5113|M.avium_104 FVSEWLLVQSLIHAAPQHNTVVALAAPLAVGAVALTTGLGVAAMVKAFGI
: * . * *: : :* .:* : *
Mb2861c|M.bovis_AF2122/97 -------DRSVLAAIGVLWWFMVVQLPFAG--IVFAVDGVLLG--AGDAA
Rv2836c|M.tuberculosis_H37Rv -------DRSVLAAIGVPWWFMVVQLPFAG--IVFAVDGVLLG--AGDAA
MMAR_1897|M.marinum_M -------DAAVLAAVTVPWWFLVAQLPFAG--IVFALDGVLLG--AGDAA
MUL_2125|M.ulcerans_Agy99 -------DAAVLAAITVPWWFLVAQLPFAG--VVFALDGVLLG--AGDAA
Mflv_4036|M.gilvum_PYR-GCK -------DQSVLDRIGVPWWFLVAQLPVAG--IVFAIDGVLLG--AGDAT
Mvan_2317|M.vanbaalenii_PYR-1 -------DRSVLDQIGVPWWFLVAQLPVAG--IVFAIDGVLLG--AGDAT
MSMEG_2631|M.smegmatis_MC2_155 -------DESVLATIGVPWWFLVAQLPVAG--IVFALDGVLLG--AGDAK
MAB_3128c|M.abscessus_ATCC_199 -------DAAVLHEMRVPWWFLVCQLPISG--LVFALDGVLLG--AADAR
MAV_5113|M.avium_104 GFLARPRSQPAADAREVPAVMVAGMALAATGCVVVAVAPLVVGSMLGRVL
. .. * ::. : :*.*: :::* . .
Mb2861c|M.bovis_AF2122/97 FMRTATVASALVGFLPLVWLSLAYG-WGLAGIWSGLGTFIVLRLIFVGWR
Rv2836c|M.tuberculosis_H37Rv FMRTATVASALVGFLPLVWLSLAYG-WGLAGIWSGLGTFIVLRLIFVGWR
MMAR_1897|M.marinum_M FMRTATVVSALAGFLPLTWLSLVYG-WGLAGIWSGLATFIVLRLIFVGWR
MUL_2125|M.ulcerans_Agy99 FMRTATVVSALAGFLPLTWLSLVYG-WGLAGIWSGLATFIVLRLIFVGWR
Mflv_4036|M.gilvum_PYR-GCK FMRNATLASALAGFLPLVWLSLAFG-WGLLGIWAGLSTFMVLRLVFVGWR
Mvan_2317|M.vanbaalenii_PYR-1 FMRNATLISALVGFLPLIWLSLAFG-WGLLGIWAGLSAFMVLRLVFVGWR
MSMEG_2631|M.smegmatis_MC2_155 FMRNATLASAMLGFLPLIWLSLAFG-WGLFGIWSGLSTFMVLRLVFVGWR
MAB_3128c|M.abscessus_ATCC_199 FMRNATMISALCGFLPSVWLALVFG-WGLAGIWCGLTLFLVLRLVLVGWR
MAV_5113|M.avium_104 TELPASRAVAFTDFGAVLRLPGLHGSIAPGAIAAALAAAVLAAVGLACWR
*: *: .* . *. .* . .* ..* :: : :. **
Mb2861c|M.bovis_AF2122/97 AYSGR-------WAVTGAA-------------------------------
Rv2836c|M.tuberculosis_H37Rv AYSGR-------WAVTGAA-------------------------------
MMAR_1897|M.marinum_M AMSGR-------WALTGTA-------------------------------
MUL_2125|M.ulcerans_Agy99 AMSGR-------WALTGTA-------------------------------
Mflv_4036|M.gilvum_PYR-GCK ALSGR-------WLVPGTR-------------------------------
Mvan_2317|M.vanbaalenii_PYR-1 ALSGR-------WLVPGTR-------------------------------
MSMEG_2631|M.smegmatis_MC2_155 AFSGR-------WLVPGTS-------------------------------
MAB_3128c|M.abscessus_ATCC_199 ALSGG-------WAVPGTGS------------------------------
MAV_5113|M.avium_104 SRRRSDPVTLPLWACGADELTPRMQYTATSFAEPLQRVFDDVLRPDTDIE
: * .
Mb2861c|M.bovis_AF2122/97 --------------------------------------------------
Rv2836c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1897|M.marinum_M --------------------------------------------------
MUL_2125|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4036|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2317|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2631|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3128c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5113|M.avium_104 ITHLTESRYLAEAVTYRTRIADAIEERLYAPVIGAVAAAAQLVRRAHTGS
Mb2861c|M.bovis_AF2122/97 --------------------
Rv2836c|M.tuberculosis_H37Rv --------------------
MMAR_1897|M.marinum_M --------------------
MUL_2125|M.ulcerans_Agy99 --------------------
Mflv_4036|M.gilvum_PYR-GCK --------------------
Mvan_2317|M.vanbaalenii_PYR-1 --------------------
MSMEG_2631|M.smegmatis_MC2_155 --------------------
MAB_3128c|M.abscessus_ATCC_199 --------------------
MAV_5113|M.avium_104 VHLYLAYGMLGVLIVLVVAR