For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLIHEHAWLQPATCGAGCVHAGADGPGRRWVVALRTTVRVVAAVTLFAGVWLLALPLPGRSHLQRGYCR LMLRSLGVRMSVSGGPIRNLRGVLVVSGHVSWLDVFAIGTVLPGSFVARADLVDWPALGPIVRIMKVIPI DRARLRRLPAVVAAVAGRLRAGYTVVAFPEGTTWCGLGYGRFRPAMFQAAIDAGRPVQPLRLSYRHKDGR PSTVPAFVGDDTLLTSIKRVITARRTVCHIHVESLQLPGVDRRDLAARCEAAVRGDAGSSARRAISGCGG AGFAARGGNHTLVA
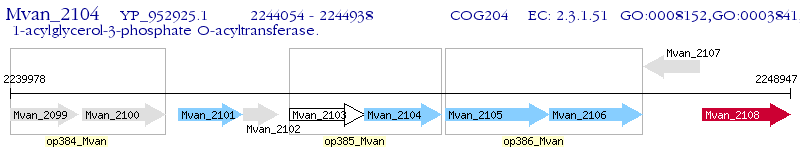
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2104 | - | - | 100% (294) | phospholipid/glycerol acyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5656 | - | 7e-05 | 27.46% (142) | phospholipid/glycerol acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3052c | - | 8e-93 | 60.00% (290) | hypothetical protein Mb3052c |
| M. gilvum PYR-GCK | Mflv_4256 | - | 1e-138 | 81.97% (294) | phospholipid/glycerol acyltransferase |
| M. tuberculosis H37Rv | Rv3026c | - | 8e-93 | 60.00% (290) | hypothetical protein Rv3026c |
| M. leprae Br4923 | MLBr_00087 | - | 3e-05 | 30.77% (117) | putative acyltransferase |
| M. abscessus ATCC 19977 | MAB_3358c | - | 1e-97 | 67.43% (261) | putative acyltransferase |
| M. marinum M | MMAR_1687 | - | 5e-93 | 62.55% (267) | hypothetical protein MMAR_1687 |
| M. avium 104 | MAV_3873 | - | 7e-86 | 59.11% (291) | 1-acylglycerol-3-phosphate O-acyltransferase |
| M. smegmatis MC2 155 | MSMEG_2356 | - | 1e-112 | 75.77% (260) | acyltransferase |
| M. thermoresistible (build 8) | TH_0895 | - | 1e-05 | 28.17% (142) | POSSIBLE ACYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_1925 | - | 4e-93 | 62.55% (267) | hypothetical protein MUL_1925 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3052c|M.bovis_AF2122/97 MSAPAVTEHSWLPRATCGVSCVSVGDAAQVRRPLVVLRVALRVMLALLLV
Rv3026c|M.tuberculosis_H37Rv MSAPAVTEHSWLPRATCGVSCVSVGDAAQVRRPLVVLRVALRVMLALLLV
MAV_3873|M.avium_104 ---------------------MAAGRPAAERRLRVVVRVTLRLLLAVLLA
MMAR_1687|M.marinum_M --MNTEAGHSWLPRTSCDISCVSLGHPAVPRRVPATLRVALRIVWALLLM
MUL_1925|M.ulcerans_Agy99 --MNTEAGHSWLPRTSCDISCVSLGHPAVPRRVPATLRVALRIVWALLLM
Mvan_2104|M.vanbaalenii_PYR-1 --MTLIHEHAWLQPATCGAGCVHAGADGPGRRWVVALRTTVRVVAAVTLF
Mflv_4256|M.gilvum_PYR-GCK --MTVTDGHAWLQAATCGAGCVDAGADGPGRRWVVALRTGVRVSAALTLV
MSMEG_2356|M.smegmatis_MC2_155 ---MTTREHPWLPKASCDAGCVHSVEQ--GRRVIVWLRTAARLSAALALL
MAB_3358c|M.abscessus_ATCC_199 --MAPEYAHAWLPKATCDESCMRANLIDEPGRFTRTFRTTSRIAMAITLL
MLBr_00087|M.leprae_Br4923 ----MAVEPLYGTVIQFARLLWLIQGLKVSVRGIENLPASGGAVIAINHT
TH_0895|M.thermoresistible__bu ------VEPVYATVIQTARLVWRLQGLTFTVTGVENLPTTGGAVIAINHT
. . *:
Mb3052c|M.bovis_AF2122/97 PGVPLVVMPLPGRTRVQRIYCRLVLRLFGVRITVSGSPVRNLRGVLVVSG
Rv3026c|M.tuberculosis_H37Rv PGVPLVVMPLPGRTRVQRIYCRLVLRLFGVRITVSGSPVRNLRGVLVVSG
MAV_3873|M.avium_104 PGWPLLAVPLPGRTRVQRIYCRLVLRCFGVRITVSGNPIRNLRGVLVVSP
MMAR_1687|M.marinum_M PGLPLLGVPVPGRSHAQRAYCRLVLRCFGVRITVSGSPIRNLRGVLVVSN
MUL_1925|M.ulcerans_Agy99 PGLPLLGVPVPGRSHAQRAYCRLVLRCFGVRITVSGSPIRNLRGVLVVSN
Mvan_2104|M.vanbaalenii_PYR-1 AGVWLLALPLPGRSHLQRGYCRLMLRSLGVRMSVSGGPIRNLRGVLVVSG
Mflv_4256|M.gilvum_PYR-GCK TGVWLLALPLPGRSHLQRGYCRLMLRSLGVRMSVAGGPVRNLRGVLVVSG
MSMEG_2356|M.smegmatis_MC2_155 PGMPLLAVPLPGRSHVQRIYCRLLLRCLGVRIRLSGGPIRNLQGVLVVSG
MAB_3358c|M.abscessus_ATCC_199 TLAPLLGIPLPGRVRWQRGYCRLMLRCLGVRISVSGGPIRDIRGSLVVAG
MLBr_00087|M.leprae_Br4923 SYLDFTFAGLPAYQQGLGRKVRFMAKQEVFDGKITGPIMRRLRHIPVDRQ
TH_0895|M.thermoresistible__bu SYFDFTFAGLPAYQQGLGRKVRFMAKKEVFDHKIGGPLMRSLRHIPVDRD
. : :*. : *:: : . : * :* :: *
Mb3052c|M.bovis_AF2122/97 HVSWLDVFCIGSVLPGSFVARADMFTGRTIGIVARILKIIPIERASLRRL
Rv3026c|M.tuberculosis_H37Rv HVSWLDVFCIGSVLPGSFVARADMFTGRTIGIVARILKIIPIERASLRRL
MAV_3873|M.avium_104 HTSWLDVFAIGAVLPGSFVARADMFSGPATGVVARILKIIPIERSSLRRL
MMAR_1687|M.marinum_M HISWLDVFAIGAVLPGSFVARADMFTGPGVEIVARLLKIIPIERTSLRRL
MUL_1925|M.ulcerans_Agy99 HISWLDVFAIGAVLPGSFVARADMFTGPGVEIVARLLKIIPIERTSLRRL
Mvan_2104|M.vanbaalenii_PYR-1 HVSWLDVFAIGTVLPGSFVARADLVDWPALGPIVRIMKVIPIDRARLRRL
Mflv_4256|M.gilvum_PYR-GCK HVSWLDVFAIGTVLPGSFVARADLVTWPALGPVVRLMKVIPIDRARLRRL
MSMEG_2356|M.smegmatis_MC2_155 HVSWLDIFAIGSVLPGSFVARADLVEWPALGRVARLMKVIPIDRASLRRL
MAB_3358c|M.abscessus_ATCC_199 HVSWVDVFAIGAVLPGSFVARADLIDWPGLGVIARMMRVIPIERGNLRTL
MLBr_00087|M.leprae_Br4923 EGAASYEAAVRNLKDGELVG------VYPEATISRSFEIKECKGGAARMS
TH_0895|M.thermoresistible__bu SGADSFAAACDALRAGELVG------VYPEATISRSFEIKELKSGAARMA
: . : *.:*. : * :.: . *
Mb3052c|M.bovis_AF2122/97 PGVVDTIARRLRAGQTVVAFPEGTTWCGR--------------PGDDAGR
Rv3026c|M.tuberculosis_H37Rv PGVVDTIARRLRAGQTVVAFPEGTTWCGR--------------PGDDAGR
MAV_3873|M.avium_104 PGVVDAVAHRLRAGQTVVAFPEGTTWCGRDGDDAGGGGGPSHTSGDDAGR
MMAR_1687|M.marinum_M PGVVDTVARRLRAGQTVVAFPEGTTWCGL---------------------
MUL_1925|M.ulcerans_Agy99 PGVVDTVARRLRAGQTVVAFPEGTTWCGL---------------------
Mvan_2104|M.vanbaalenii_PYR-1 PAVVAAVAGRLRAGYTVVAFPEGTTWCGL---------------------
Mflv_4256|M.gilvum_PYR-GCK PTVVASIADRLRAGQTVVAFPEGTTWCGL---------------------
MSMEG_2356|M.smegmatis_MC2_155 PAVVATVADRLRAGRTVVAFPEGTTWCGL---------------------
MAB_3358c|M.abscessus_ATCC_199 PRVVGTVAARLRAGQTVVAFPEGTTWCGR---------------------
MLBr_00087|M.leprae_Br4923 AEARVPIIP-------HIVWGSQRIWTKD---------------------
TH_0895|M.thermoresistible__bu IECGVPIVP-------HIVWGAQRIWTKG---------------------
.: :.: *
Mb3052c|M.bovis_AF2122/97 PAARAGAGCSHRGCGAFYPAMFQAAIDAGRPVQPLRLTYHHVDGTVSTAP
Rv3026c|M.tuberculosis_H37Rv PAARAGAGCSHRGCGAFYPAMFQAAIDAGRPVQPLRLTYHHVDGTVSTAP
MAV_3873|M.avium_104 GAAGGGGGPSHRGRGAFYPAMFQAAIDAGRPVQPLRLTYQHVDGSTSTAP
MMAR_1687|M.marinum_M ------------ASGSFYPAMFQAAIDAGRPVQPLRLTYHHVNGAISTTP
MUL_1925|M.ulcerans_Agy99 ------------ASGSFYPAMFQAAIDAGRPVQPLRLTYHHVNGAISTTP
Mvan_2104|M.vanbaalenii_PYR-1 ------------GYGRFRPAMFQAAIDAGRPVQPLRLSYRHKDGRPSTVP
Mflv_4256|M.gilvum_PYR-GCK ------------GYGRFRPAMFQAAVDAGRPVQPLRVSYRHRDGRPSTVP
MSMEG_2356|M.smegmatis_MC2_155 ------------AHGPFRPAMFQAAVDAARPVQPLRLTYRHRDGRPSTTP
MAB_3358c|M.abscessus_ATCC_199 ------------AYGSFRPAMFQAAVDAQRPVQPLRLTYHHLNGELSTVP
MLBr_00087|M.leprae_Br4923 ------------CPKKLLRPKVPILVLVGEPIEPT-LGVAQLKALLHFRM
TH_0895|M.thermoresistible__bu ------------HPRKLLRPKVPISVAVGEPIQPT-LPAPELTALLRSRM
: . . : . .*::* : . .
Mb3052c|M.bovis_AF2122/97 AFVGDDTLVRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLA
Rv3026c|M.tuberculosis_H37Rv AFVGDDTLVRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLA
MAV_3873|M.avium_104 AFVGDETLLGSIRRLLTVRRTLARVQVESLQLPGTDRRALAGRCQSAVGV
MMAR_1687|M.marinum_M AFVGDDTLARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHA
MUL_1925|M.ulcerans_Agy99 AFVGDDTLARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHA
Mvan_2104|M.vanbaalenii_PYR-1 AFVGDDTLLTSIKRVITARRTVCHIHVESLQLPGVDRRDLAARCEAAVRG
Mflv_4256|M.gilvum_PYR-GCK AFVGDDSLLDSITRVITARRTVCHISVESLQLPGRDRRELAARCESAVRG
MSMEG_2356|M.smegmatis_MC2_155 AFVGQDTLLRSVCRVVTARRTVCHVHVGALQLPGDDRRALAARCEAEVRG
MAB_3358c|M.abscessus_ATCC_199 AYVGEDTLMASIRRLTATRMTVAHIRVAGLQLPGESRRDLAARCETAVRA
MLBr_00087|M.leprae_Br4923 QHLLERAQELYGPHPAGEFWVPHRLGGGAPSLIKAARLDAEEATERAARR
TH_0895|M.thermoresistible__bu QHLLEQVQDSYGPHPPGEFWVPKRLGGGAPSLAEADQMDVEEARQKAARR
.: : : : . .* : : .
Mb3052c|M.bovis_AF2122/97 GALGQSGQRPG-------------RRHVPAT
Rv3026c|M.tuberculosis_H37Rv GALGQSGQRPG-------------RRHVPAT
MAV_3873|M.avium_104 AAARRAG--HG-------------RVLVA--
MMAR_1687|M.marinum_M GAAHPAHR----------------PALVA--
MUL_1925|M.ulcerans_Agy99 GAAHPAHR----------------PALVA--
Mvan_2104|M.vanbaalenii_PYR-1 DAGSSARRAISGCGGAGFAARGGNHTLVA--
Mflv_4256|M.gilvum_PYR-GCK HSAPVPRATAAACGGG--------HALVA--
MSMEG_2356|M.smegmatis_MC2_155 --GHERRDART-------------HAIAA--
MAB_3358c|M.abscessus_ATCC_199 GDQIPQVHSHHHPEHPE-----RPEPIAA--
MLBr_00087|M.leprae_Br4923 AQR----AKVAE-------------------
TH_0895|M.thermoresistible__bu AARDAEGAGTSESPG----------------