For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVTDGHAWLQAATCGAGCVDAGADGPGRRWVVALRTGVRVSAALTLVTGVWLLALPLPGRSHLQRGYCRL MLRSLGVRMSVAGGPVRNLRGVLVVSGHVSWLDVFAIGTVLPGSFVARADLVTWPALGPVVRLMKVIPID RARLRRLPTVVASIADRLRAGQTVVAFPEGTTWCGLGYGRFRPAMFQAAVDAGRPVQPLRVSYRHRDGRP STVPAFVGDDSLLDSITRVITARRTVCHISVESLQLPGRDRRELAARCESAVRGHSAPVPRATAAACGGG HALVA*
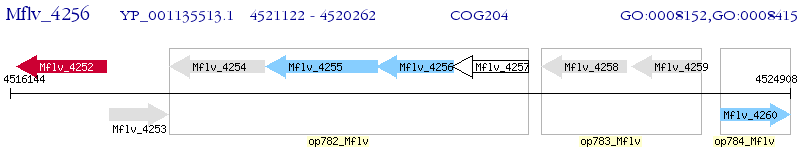
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4256 | - | - | 100% (286) | phospholipid/glycerol acyltransferase |
| M. gilvum PYR-GCK | Mflv_2564 | - | 7e-06 | 21.13% (265) | HAD family hydrolase |
| M. gilvum PYR-GCK | Mflv_1153 | - | 6e-05 | 28.28% (145) | phospholipid/glycerol acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3052c | - | 2e-92 | 62.32% (276) | hypothetical protein Mb3052c |
| M. tuberculosis H37Rv | Rv3026c | - | 2e-92 | 62.32% (276) | hypothetical protein Rv3026c |
| M. leprae Br4923 | MLBr_01245 | - | 4e-08 | 26.01% (173) | putative transferase |
| M. abscessus ATCC 19977 | MAB_3358c | - | 7e-95 | 65.64% (259) | putative acyltransferase |
| M. marinum M | MMAR_1687 | - | 9e-93 | 62.74% (263) | hypothetical protein MMAR_1687 |
| M. avium 104 | MAV_3873 | - | 2e-83 | 55.67% (300) | 1-acylglycerol-3-phosphate O-acyltransferase |
| M. smegmatis MC2 155 | MSMEG_2356 | - | 1e-110 | 74.43% (262) | acyltransferase |
| M. thermoresistible (build 8) | TH_0895 | - | 1e-05 | 27.46% (142) | POSSIBLE ACYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_1925 | - | 6e-93 | 62.74% (263) | hypothetical protein MUL_1925 |
| M. vanbaalenii PYR-1 | Mvan_2104 | - | 1e-138 | 81.97% (294) | phospholipid/glycerol acyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 MSAFDENGTPKSGPEVRLPGSVAEILASPAGPQVGAFFDLDGTLVAGFTA
TH_0895|M.thermoresistible__bu --------------------------------------------------
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 VILTHERLRRCDMGVGELLSMIQAGLNHTLGRIEFEDLIGKVSSALRGRL
TH_0895|M.thermoresistible__bu --------------------------------------------------
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 LTDLEEIGERLFAQRIESRIYPEMRKLVQAHVARGHTVVLSSSALTIQVG
TH_0895|M.thermoresistible__bu --------------------------------------------------
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 PVARFLGIAHMLTNKFEINEDGMLTGGVVKPILWGPGKAAAVQRFAAEHD
TH_0895|M.thermoresistible__bu --------------------------------------------------
Mflv_4256|M.gilvum_PYR-GCK -------------------------MTVTDGHAWLQAATCGAGCVDAGAD
Mvan_2104|M.vanbaalenii_PYR-1 -------------------------MTLIHEHAWLQPATCGAGCVHAGAD
MSMEG_2356|M.smegmatis_MC2_155 --------------------------MTTREHPWLPKASCDAGCVHSVEQ
MAB_3358c|M.abscessus_ATCC_199 -------------------------MAPEYAHAWLPKATCDESCMRANLI
Mb3052c|M.bovis_AF2122/97 -----------------------MSAPAVTEHSWLPRATCGVSCVSVGDA
Rv3026c|M.tuberculosis_H37Rv -----------------------MSAPAVTEHSWLPRATCGVSCVSVGDA
MAV_3873|M.avium_104 --------------------------------------------MAAGRP
MMAR_1687|M.marinum_M -------------------------MNTEAGHSWLPRTSCDISCVSLGHP
MUL_1925|M.ulcerans_Agy99 -------------------------MNTEAGHSWLPRTSCDISCVSLGHP
MLBr_01245|M.leprae_Br4923 IDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPILKFNS
TH_0895|M.thermoresistible__bu -----------------------------VEPVYATVIQTARLVWRLQGL
Mflv_4256|M.gilvum_PYR-GCK GPGRRWVVALRTGVRVSAALTLVTGVWLLALPLPGRSH----LQRGYCRL
Mvan_2104|M.vanbaalenii_PYR-1 GPGRRWVVALRTTVRVVAAVTLFAGVWLLALPLPGRSH----LQRGYCRL
MSMEG_2356|M.smegmatis_MC2_155 --GRRVIVWLRTAARLSAALALLPGMPLLAVPLPGRSH----VQRIYCRL
MAB_3358c|M.abscessus_ATCC_199 DEPGRFTRTFRTTSRIAMAITLLTLAPLLGIPLPGRVR----WQRGYCRL
Mb3052c|M.bovis_AF2122/97 AQVRRPLVVLRVALRVMLALLLVPGVPLVVMPLPGRTR----VQRIYCRL
Rv3026c|M.tuberculosis_H37Rv AQVRRPLVVLRVALRVMLALLLVPGVPLVVMPLPGRTR----VQRIYCRL
MAV_3873|M.avium_104 AAERRLRVVVRVTLRLLLAVLLAPGWPLLAVPLPGRTR----VQRIYCRL
MMAR_1687|M.marinum_M AVPRRVPATLRVALRIVWALLLMPGLPLLGVPVPGRSH----AQRAYCRL
MUL_1925|M.ulcerans_Agy99 AVPRRVPATLRVALRIVWALLLMPGLPLLGVPVPGRSH----AQRAYCRL
MLBr_01245|M.leprae_Br4923 RGGVGIGAQLRTLVGLSSVLPLVAGAVGIGVLTGSRRRGANFFTSVFPPL
TH_0895|M.thermoresistible__bu TFTVTGVENLPTTGGAVIAINHTSYFDFTFAGLPAYQQG-----LGRKVR
. . .: . . :
Mflv_4256|M.gilvum_PYR-GCK MLRSLGVRMSVAGGP-VRNLRGVLVVSGHVSWLDVFAIGTVLPG--SFVA
Mvan_2104|M.vanbaalenii_PYR-1 MLRSLGVRMSVSGGP-IRNLRGVLVVSGHVSWLDVFAIGTVLPG--SFVA
MSMEG_2356|M.smegmatis_MC2_155 LLRCLGVRIRLSGGP-IRNLQGVLVVSGHVSWLDIFAIGSVLPG--SFVA
MAB_3358c|M.abscessus_ATCC_199 MLRCLGVRISVSGGP-IRDIRGSLVVAGHVSWVDVFAIGAVLPG--SFVA
Mb3052c|M.bovis_AF2122/97 VLRLFGVRITVSGSP-VRNLRGVLVVSGHVSWLDVFCIGSVLPG--SFVA
Rv3026c|M.tuberculosis_H37Rv VLRLFGVRITVSGSP-VRNLRGVLVVSGHVSWLDVFCIGSVLPG--SFVA
MAV_3873|M.avium_104 VLRCFGVRITVSGNP-IRNLRGVLVVSPHTSWLDVFAIGAVLPG--SFVA
MMAR_1687|M.marinum_M VLRCFGVRITVSGSP-IRNLRGVLVVSNHISWLDVFAIGAVLPG--SFVA
MUL_1925|M.ulcerans_Agy99 VLRCFGVRITVSGSP-IRNLRGVLVVSNHISWLDVFAIGAVLPG--SFVA
MLBr_01245|M.leprae_Br4923 VLAAAGVHLNVIGNENLTMQRPAVFIYNHRNQIDPLIAGALVRGNWIAVA
TH_0895|M.thermoresistible__bu FMAKKEVFDHKIGGPLMRSLRHIPVDRDSGADSFAAACDALRAG------
.: * *. : : . .:: *
Mflv_4256|M.gilvum_PYR-GCK RADLVTWPALGPVVRLMKVIPIDRARLRRLPTVVASIADRLRAGQTVVAF
Mvan_2104|M.vanbaalenii_PYR-1 RADLVDWPALGPIVRIMKVIPIDRARLRRLPAVVAAVAGRLRAGYTVVAF
MSMEG_2356|M.smegmatis_MC2_155 RADLVEWPALGRVARLMKVIPIDRASLRRLPAVVATVADRLRAGRTVVAF
MAB_3358c|M.abscessus_ATCC_199 RADLIDWPGLGVIARMMRVIPIERGNLRTLPRVVGTVAARLRAGQTVVAF
Mb3052c|M.bovis_AF2122/97 RADMFTGRTIGIVARILKIIPIERASLRRLPGVVDTIARRLRAGQTVVAF
Rv3026c|M.tuberculosis_H37Rv RADMFTGRTIGIVARILKIIPIERASLRRLPGVVDTIARRLRAGQTVVAF
MAV_3873|M.avium_104 RADMFSGPATGVVARILKIIPIERSSLRRLPGVVDAVAHRLRAGQTVVAF
MMAR_1687|M.marinum_M RADMFTGPGVEIVARLLKIIPIERTSLRRLPGVVDTVARRLRAGQTVVAF
MUL_1925|M.ulcerans_Agy99 RADMFTGPGVEIVARLLKIIPIERTSLRRLPGVVDTVARRLRAGQTVVAF
MLBr_01245|M.leprae_Br4923 KKELAKNPIIRALGKLTDGVFIDRDDPVGAVETLHAVEDRARKGLSILMA
TH_0895|M.thermoresistible__bu --ELVGVYPEATISRSFEIKELKSGAARMAIECGVPIVPHIVWGAQRIWT
:: : : :. .: : * :
Mflv_4256|M.gilvum_PYR-GCK PEGTTWCGL---------------------------------GYGRFRPA
Mvan_2104|M.vanbaalenii_PYR-1 PEGTTWCGL---------------------------------GYGRFRPA
MSMEG_2356|M.smegmatis_MC2_155 PEGTTWCGL---------------------------------AHGPFRPA
MAB_3358c|M.abscessus_ATCC_199 PEGTTWCGR---------------------------------AYGSFRPA
Mb3052c|M.bovis_AF2122/97 PEGTTWCGR--------------PGDDAGRPAARAGAGCSHRGCGAFYPA
Rv3026c|M.tuberculosis_H37Rv PEGTTWCGR--------------PGDDAGRPAARAGAGCSHRGCGAFYPA
MAV_3873|M.avium_104 PEGTTWCGRDGDDAGGGGGPSHTSGDDAGRGAAGGGGGPSHRGRGAFYPA
MMAR_1687|M.marinum_M PEGTTWCGL---------------------------------ASGSFYPA
MUL_1925|M.ulcerans_Agy99 PEGTTWCGL---------------------------------ASGSFYPA
MLBr_01245|M.leprae_Br4923 PEGTRLDTT---------------------------------EVGPFKKG
TH_0895|M.thermoresistible__bu KG----------------------------------------HPRKLLRP
:
Mflv_4256|M.gilvum_PYR-GCK MFQAAVDAGRPVQPLRVSYRHRDGR------------PSTVPAFVGDDSL
Mvan_2104|M.vanbaalenii_PYR-1 MFQAAIDAGRPVQPLRLSYRHKDGR------------PSTVPAFVGDDTL
MSMEG_2356|M.smegmatis_MC2_155 MFQAAVDAARPVQPLRLTYRHRDGR------------PSTTPAFVGQDTL
MAB_3358c|M.abscessus_ATCC_199 MFQAAVDAQRPVQPLRLTYHHLNGE------------LSTVPAYVGEDTL
Mb3052c|M.bovis_AF2122/97 MFQAAIDAGRPVQPLRLTYHHVDGT------------VSTAPAFVGDDTL
Rv3026c|M.tuberculosis_H37Rv MFQAAIDAGRPVQPLRLTYHHVDGT------------VSTAPAFVGDDTL
MAV_3873|M.avium_104 MFQAAIDAGRPVQPLRLTYQHVDGS------------TSTAPAFVGDETL
MMAR_1687|M.marinum_M MFQAAIDAGRPVQPLRLTYHHVNGA------------ISTTPAFVGDDTL
MUL_1925|M.ulcerans_Agy99 MFQAAIDAGRPVQPLRLTYHHVNGA------------ISTTPAFVGDDTL
MLBr_01245|M.leprae_Br4923 PFRIAMAVGIPIVPIVFRNAEIVAARNSNTINPGMVDVAVLPAIPVDDWT
TH_0895|M.thermoresistible__bu KVPISVAVGEPIQPT-LPAPELTAL------------LRSRMQHLLEQVQ
. :: . *: * . . . ::
Mflv_4256|M.gilvum_PYR-GCK LDSITRVITARRTVCHISVESLQLPGRDRRELAARCESAVRGHSAPVPRA
Mvan_2104|M.vanbaalenii_PYR-1 LTSIKRVITARRTVCHIHVESLQLPGVDRRDLAARCEAAVRGDAGSSARR
MSMEG_2356|M.smegmatis_MC2_155 LRSVCRVVTARRTVCHVHVGALQLPGDDRRALAARCEAEVRG--GHERRD
MAB_3358c|M.abscessus_ATCC_199 MASIRRLTATRMTVAHIRVAGLQLPGESRRDLAARCETAVRAGDQIPQVH
Mb3052c|M.bovis_AF2122/97 VRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLAGALGQSGQ
Rv3026c|M.tuberculosis_H37Rv VRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLAGALGQSGQ
MAV_3873|M.avium_104 LGSIRRLLTVRRTLARVQVESLQLPGTDRRALAGRCQSAVGVAAARRAG-
MMAR_1687|M.marinum_M ARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHAGAAHPAHR
MUL_1925|M.ulcerans_Agy99 ARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHAGAAHPAHR
MLBr_01245|M.leprae_Br4923 LDALSERIAEVRQLYLDILADWPVDELPKAALYTRATKATVKKAEPKKGG
TH_0895|M.thermoresistible__bu DSYGPHPPGEFWVPKRLGGGAPSLAEADQMDVEEARQKAARRAARDAEGA
. : : : .
Mflv_4256|M.gilvum_PYR-GCK TAAACGGG--------HALVA-----------------------------
Mvan_2104|M.vanbaalenii_PYR-1 AISGCGGAGFAARGGNHTLVA-----------------------------
MSMEG_2356|M.smegmatis_MC2_155 ART-------------HAIAA-----------------------------
MAB_3358c|M.abscessus_ATCC_199 SHHHPEHPE-----RPEPIAA-----------------------------
Mb3052c|M.bovis_AF2122/97 RPG-------------RRHVPAT---------------------------
Rv3026c|M.tuberculosis_H37Rv RPG-------------RRHVPAT---------------------------
MAV_3873|M.avium_104 -HG-------------RVLVA-----------------------------
MMAR_1687|M.marinum_M ----------------PALVA-----------------------------
MUL_1925|M.ulcerans_Agy99 ----------------PALVA-----------------------------
MLBr_01245|M.leprae_Br4923 RTSAAKTTAKKTATKATAAKTANTAVTKSMLTPPTRISDKENPKVPKVVS
TH_0895|M.thermoresistible__bu GTS-------------ESPG------------------------------
Mflv_4256|M.gilvum_PYR-GCK ------------
Mvan_2104|M.vanbaalenii_PYR-1 ------------
MSMEG_2356|M.smegmatis_MC2_155 ------------
MAB_3358c|M.abscessus_ATCC_199 ------------
Mb3052c|M.bovis_AF2122/97 ------------
Rv3026c|M.tuberculosis_H37Rv ------------
MAV_3873|M.avium_104 ------------
MMAR_1687|M.marinum_M ------------
MUL_1925|M.ulcerans_Agy99 ------------
MLBr_01245|M.leprae_Br4923 QDLVVKQPRERS
TH_0895|M.thermoresistible__bu ------------