For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
APEYAHAWLPKATCDESCMRANLIDEPGRFTRTFRTTSRIAMAITLLTLAPLLGIPLPGRVRWQRGYCRL MLRCLGVRISVSGGPIRDIRGSLVVAGHVSWVDVFAIGAVLPGSFVARADLIDWPGLGVIARMMRVIPIE RGNLRTLPRVVGTVAARLRAGQTVVAFPEGTTWCGRAYGSFRPAMFQAAVDAQRPVQPLRLTYHHLNGEL STVPAYVGEDTLMASIRRLTATRMTVAHIRVAGLQLPGESRRDLAARCETAVRAGDQIPQVHSHHHPEHP ERPEPIAA*
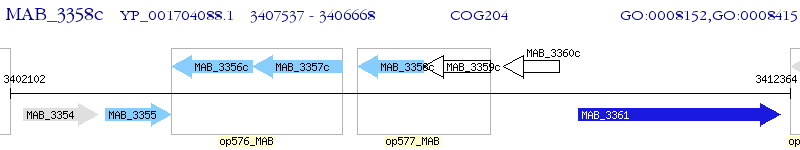
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3358c | - | - | 100% (289) | putative acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3052c | - | 3e-86 | 56.27% (279) | hypothetical protein Mb3052c |
| M. gilvum PYR-GCK | Mflv_4256 | - | 8e-95 | 65.64% (259) | phospholipid/glycerol acyltransferase |
| M. tuberculosis H37Rv | Rv3026c | - | 3e-86 | 56.27% (279) | hypothetical protein Rv3026c |
| M. leprae Br4923 | MLBr_01245 | - | 2e-05 | 24.06% (133) | putative transferase |
| M. marinum M | MMAR_1687 | - | 1e-95 | 61.15% (278) | hypothetical protein MMAR_1687 |
| M. avium 104 | MAV_3873 | - | 2e-79 | 57.47% (261) | 1-acylglycerol-3-phosphate O-acyltransferase |
| M. smegmatis MC2 155 | MSMEG_2356 | - | 5e-97 | 62.45% (269) | acyltransferase |
| M. thermoresistible (build 8) | TH_1038 | - | 2e-06 | 25.13% (187) | POSSIBLE TRANSMEMBRANE PHOSPHOLIPID BIOSYNTHESIS BIFUNCTIONNAL |
| M. ulcerans Agy99 | MUL_1925 | - | 8e-96 | 61.15% (278) | hypothetical protein MUL_1925 |
| M. vanbaalenii PYR-1 | Mvan_2104 | - | 2e-97 | 67.43% (261) | phospholipid/glycerol acyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 MSAFDENG-TPKSGPEVRLPGSVAEILASPAGPQVGAFFDLDGTLVAGFT
TH_1038|M.thermoresistible__bu MTAEDRRAERDEEERTLRLPGSVAEIEASPPGPEIGAFFDLDGTLVAGFT
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 AVILTHERLRRCDMGVGELLSMIQAGLNHTLGRIEFEDLIGKVSSALRGR
TH_1038|M.thermoresistible__bu GVIMTRERLRRRELSVGEFIGLVQAGLNHQLGRAEFEDLIGRGARMLRGS
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 LLTDLEEIGERLFAQRIESRIYPEMRKLVQAHVARGHTVVLSSSALTIQV
TH_1038|M.thermoresistible__bu SLSDIDELAERLFVQKIQNRIYPEMRELVRAHQARGHTVVLSSSALTVQV
Mflv_4256|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2104|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2356|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3358c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3052c|M.bovis_AF2122/97 --------------------------------------------------
Rv3026c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3873|M.avium_104 --------------------------------------------------
MMAR_1687|M.marinum_M --------------------------------------------------
MUL_1925|M.ulcerans_Agy99 --------------------------------------------------
MLBr_01245|M.leprae_Br4923 GPVARFLGIAHMLTNKFEINEDGMLTGGVVKPILWGPGKAAAVQRFAAEH
TH_1038|M.thermoresistible__bu EPVARYLGIENILSNKFEVDENGCITGEVNRPIIWGPGKARAVQAFAAER
Mflv_4256|M.gilvum_PYR-GCK ------------------MTVTDGHAWLQAATCGAGCVDAGADGPGRRWV
Mvan_2104|M.vanbaalenii_PYR-1 ------------------MTLIHEHAWLQPATCGAGCVHAGADGPGRRWV
MSMEG_2356|M.smegmatis_MC2_155 -------------------MTTREHPWLPKASCDAGCVHSVEQ--GRRVI
MAB_3358c|M.abscessus_ATCC_199 ------------------MAPEYAHAWLPKATCDESCMRANLIDEPGRFT
Mb3052c|M.bovis_AF2122/97 ----------------MSAPAVTEHSWLPRATCGVSCVSVGDAAQVRRPL
Rv3026c|M.tuberculosis_H37Rv ----------------MSAPAVTEHSWLPRATCGVSCVSVGDAAQVRRPL
MAV_3873|M.avium_104 -------------------------------------MAAGRPAAERRLR
MMAR_1687|M.marinum_M ------------------MNTEAGHSWLPRTSCDISCVSLGHPAVPRRVP
MUL_1925|M.ulcerans_Agy99 ------------------MNTEAGHSWLPRTSCDISCVSLGHPAVPRRVP
MLBr_01245|M.leprae_Br4923 DIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPILKFN
TH_1038|M.thermoresistible__bu GIDLSRSYFYADGDEDVALMYLVGNPRPTNPAGKMASVAAKRGWPILRFT
: :
Mflv_4256|M.gilvum_PYR-GCK --------VALRTGVRVSAALTLVTGVWLLALPLPGR----SHLQRGYCR
Mvan_2104|M.vanbaalenii_PYR-1 --------VALRTTVRVVAAVTLFAGVWLLALPLPGR----SHLQRGYCR
MSMEG_2356|M.smegmatis_MC2_155 --------VWLRTAARLSAALALLPGMPLLAVPLPGR----SHVQRIYCR
MAB_3358c|M.abscessus_ATCC_199 --------RTFRTTSRIAMAITLLTLAPLLGIPLPGR----VRWQRGYCR
Mb3052c|M.bovis_AF2122/97 --------VVLRVALRVMLALLLVPGVPLVVMPLPGR----TRVQRIYCR
Rv3026c|M.tuberculosis_H37Rv --------VVLRVALRVMLALLLVPGVPLVVMPLPGR----TRVQRIYCR
MAV_3873|M.avium_104 --------VVVRVTLRLLLAVLLAPGWPLLAVPLPGR----TRVQRIYCR
MMAR_1687|M.marinum_M --------ATLRVALRIVWALLLMPGLPLLGVPVPGR----SHAQRAYCR
MUL_1925|M.ulcerans_Agy99 --------ATLRVALRIVWALLLMPGLPLLGVPVPGR----SHAQRAYCR
MLBr_01245|M.leprae_Br4923 SRGGVGIGAQLRTLVGLSSVLPLVAGAVGIGVLTGSRRRGANFFTSVFPP
TH_1038|M.thermoresistible__bu SRRGSNPVSQVRTAVGVASLLPVAAGAVGVGLLSRSRRAGVNFLTSVWGK
.*. : : : . : : .* :
Mflv_4256|M.gilvum_PYR-GCK LMLRSLGVRMSVAGGP-VRNLRGVLVVSGHVSWLDVFAIGTVLPG--SFV
Mvan_2104|M.vanbaalenii_PYR-1 LMLRSLGVRMSVSGGP-IRNLRGVLVVSGHVSWLDVFAIGTVLPG--SFV
MSMEG_2356|M.smegmatis_MC2_155 LLLRCLGVRIRLSGGP-IRNLQGVLVVSGHVSWLDIFAIGSVLPG--SFV
MAB_3358c|M.abscessus_ATCC_199 LMLRCLGVRISVSGGP-IRDIRGSLVVAGHVSWVDVFAIGAVLPG--SFV
Mb3052c|M.bovis_AF2122/97 LVLRLFGVRITVSGSP-VRNLRGVLVVSGHVSWLDVFCIGSVLPG--SFV
Rv3026c|M.tuberculosis_H37Rv LVLRLFGVRITVSGSP-VRNLRGVLVVSGHVSWLDVFCIGSVLPG--SFV
MAV_3873|M.avium_104 LVLRCFGVRITVSGNP-IRNLRGVLVVSPHTSWLDVFAIGAVLPG--SFV
MMAR_1687|M.marinum_M LVLRCFGVRITVSGSP-IRNLRGVLVVSNHISWLDVFAIGAVLPG--SFV
MUL_1925|M.ulcerans_Agy99 LVLRCFGVRITVSGSP-IRNLRGVLVVSNHISWLDVFAIGAVLPG--SFV
MLBr_01245|M.leprae_Br4923 LVLAAAGVHLNVIGNENLTMQRPAVFIYNHRNQIDPLIAGALVRGNWIAV
TH_1038|M.thermoresistible__bu LLLAATGVELNVLGEENLTAERPAVFIFNHRNQVDPVIAGRLVKDNFTMV
*:* **.: : * : : :.: * . :* . * :: . *
Mflv_4256|M.gilvum_PYR-GCK ARADLVTWPALGPVVRLMKVIPIDRARLRRLPTVVASIADRLRAGQTVVA
Mvan_2104|M.vanbaalenii_PYR-1 ARADLVDWPALGPIVRIMKVIPIDRARLRRLPAVVAAVAGRLRAGYTVVA
MSMEG_2356|M.smegmatis_MC2_155 ARADLVEWPALGRVARLMKVIPIDRASLRRLPAVVATVADRLRAGRTVVA
MAB_3358c|M.abscessus_ATCC_199 ARADLIDWPGLGVIARMMRVIPIERGNLRTLPRVVGTVAARLRAGQTVVA
Mb3052c|M.bovis_AF2122/97 ARADMFTGRTIGIVARILKIIPIERASLRRLPGVVDTIARRLRAGQTVVA
Rv3026c|M.tuberculosis_H37Rv ARADMFTGRTIGIVARILKIIPIERASLRRLPGVVDTIARRLRAGQTVVA
MAV_3873|M.avium_104 ARADMFSGPATGVVARILKIIPIERSSLRRLPGVVDAVAHRLRAGQTVVA
MMAR_1687|M.marinum_M ARADMFTGPGVEIVARLLKIIPIERTSLRRLPGVVDTVARRLRAGQTVVA
MUL_1925|M.ulcerans_Agy99 ARADMFTGPGVEIVARLLKIIPIERTSLRRLPGVVDTVARRLRAGQTVVA
MLBr_01245|M.leprae_Br4923 AKKELAKNPIIRALGKLTDGVFIDRDDPVGAVETLHAVEDRARKGLSILM
TH_1038|M.thermoresistible__bu GKKELADDPLLGMLGKVMDVAFIDRSDPTAAVEGLRKVEELAAKGVSILI
.: :: : :: *:* : : * :::
Mflv_4256|M.gilvum_PYR-GCK FPEGTTWCGL---------------------------------GYGRFRP
Mvan_2104|M.vanbaalenii_PYR-1 FPEGTTWCGL---------------------------------GYGRFRP
MSMEG_2356|M.smegmatis_MC2_155 FPEGTTWCGL---------------------------------AHGPFRP
MAB_3358c|M.abscessus_ATCC_199 FPEGTTWCGR---------------------------------AYGSFRP
Mb3052c|M.bovis_AF2122/97 FPEGTTWCGR--------------PGDDAGRPAARAGAGCSHRGCGAFYP
Rv3026c|M.tuberculosis_H37Rv FPEGTTWCGR--------------PGDDAGRPAARAGAGCSHRGCGAFYP
MAV_3873|M.avium_104 FPEGTTWCGRDGDDAGGGGGPSHTSGDDAGRGAAGGGGGPSHRGRGAFYP
MMAR_1687|M.marinum_M FPEGTTWCGL---------------------------------ASGSFYP
MUL_1925|M.ulcerans_Agy99 FPEGTTWCGL---------------------------------ASGSFYP
MLBr_01245|M.leprae_Br4923 APEGTRLDTT---------------------------------EVGPFKK
TH_1038|M.thermoresistible__bu APEGTRVDTN---------------------------------EVGPFKK
**** * *
Mflv_4256|M.gilvum_PYR-GCK AMFQAAVDAGRPVQPLRVSYRHRDGR------------PSTVPAFVGDDS
Mvan_2104|M.vanbaalenii_PYR-1 AMFQAAIDAGRPVQPLRLSYRHKDGR------------PSTVPAFVGDDT
MSMEG_2356|M.smegmatis_MC2_155 AMFQAAVDAARPVQPLRLTYRHRDGR------------PSTTPAFVGQDT
MAB_3358c|M.abscessus_ATCC_199 AMFQAAVDAQRPVQPLRLTYHHLNGE------------LSTVPAYVGEDT
Mb3052c|M.bovis_AF2122/97 AMFQAAIDAGRPVQPLRLTYHHVDGT------------VSTAPAFVGDDT
Rv3026c|M.tuberculosis_H37Rv AMFQAAIDAGRPVQPLRLTYHHVDGT------------VSTAPAFVGDDT
MAV_3873|M.avium_104 AMFQAAIDAGRPVQPLRLTYQHVDGS------------TSTAPAFVGDET
MMAR_1687|M.marinum_M AMFQAAIDAGRPVQPLRLTYHHVNGA------------ISTTPAFVGDDT
MUL_1925|M.ulcerans_Agy99 AMFQAAIDAGRPVQPLRLTYHHVNGA------------ISTTPAFVGDDT
MLBr_01245|M.leprae_Br4923 GPFRIAMAVGIPIVPIVFRNAEIVAARNSNTINPGMVDVAVLPAIPVDDW
TH_1038|M.thermoresistible__bu GAFRIAMATGIPIVPIVIRNAEVVAARNSTTFNPGKVDVVVYPPIPVDDW
. *: *: . *: *: . . . . *. ::
Mflv_4256|M.gilvum_PYR-GCK LLDSITRVITARRTVCHISVESLQLPGRDRRELAARCESAVRGHSAPVPR
Mvan_2104|M.vanbaalenii_PYR-1 LLTSIKRVITARRTVCHIHVESLQLPGVDRRDLAARCEAAVRGDAGSSAR
MSMEG_2356|M.smegmatis_MC2_155 LLRSVCRVVTARRTVCHVHVGALQLPGDDRRALAARCEAEVRG--GHERR
MAB_3358c|M.abscessus_ATCC_199 LMASIRRLTATRMTVAHIRVAGLQLPGESRRDLAARCETAVRAGDQIPQV
Mb3052c|M.bovis_AF2122/97 LVRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLAGALGQSG
Rv3026c|M.tuberculosis_H37Rv LVRSVCRLLTVRRTLAWVRVESLQLPGTDRRNLARRCQSAVLAGALGQSG
MAV_3873|M.avium_104 LLGSIRRLLTVRRTLARVQVESLQLPGTDRRALAGRCQSAVGVAAARRAG
MMAR_1687|M.marinum_M LARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHAGAAHPAH
MUL_1925|M.ulcerans_Agy99 LARSVRRVLAVRRIVASVHVDSLQLPGRDRRALASRCQSAVHAGAAHPAH
MLBr_01245|M.leprae_Br4923 TLDALSERIAEVRQLYLDILADWPVDELPKAALYTRATKATVKKAEPKKG
TH_1038|M.thermoresistible__bu TVEELPERIAEVRQLYLDTLKAWPRKHLPLPELYTRP-RGPAKKAPVKK-
: . : : : * *
Mflv_4256|M.gilvum_PYR-GCK ATAAACGGG--------HALVA----------------------------
Mvan_2104|M.vanbaalenii_PYR-1 RAISGCGGAGFAARGGNHTLVA----------------------------
MSMEG_2356|M.smegmatis_MC2_155 DART-------------HAIAA----------------------------
MAB_3358c|M.abscessus_ATCC_199 HSHHHPEHPE-----RPEPIAA----------------------------
Mb3052c|M.bovis_AF2122/97 QRPG-------------RRHVPAT--------------------------
Rv3026c|M.tuberculosis_H37Rv QRPG-------------RRHVPAT--------------------------
MAV_3873|M.avium_104 --HG-------------RVLVA----------------------------
MMAR_1687|M.marinum_M R----------------PALVA----------------------------
MUL_1925|M.ulcerans_Agy99 R----------------PALVA----------------------------
MLBr_01245|M.leprae_Br4923 GRTSAAKTTAKKTATKATAAKTANTAVTKSMLTPPTRISDKENPKVPKVV
TH_1038|M.thermoresistible__bu --SQAKKTQAKKTPVKKTQAKKAQAKKAQ---------AGKSADHPPGLG
Mflv_4256|M.gilvum_PYR-GCK -------------
Mvan_2104|M.vanbaalenii_PYR-1 -------------
MSMEG_2356|M.smegmatis_MC2_155 -------------
MAB_3358c|M.abscessus_ATCC_199 -------------
Mb3052c|M.bovis_AF2122/97 -------------
Rv3026c|M.tuberculosis_H37Rv -------------
MAV_3873|M.avium_104 -------------
MMAR_1687|M.marinum_M -------------
MUL_1925|M.ulcerans_Agy99 -------------
MLBr_01245|M.leprae_Br4923 SQDLVVKQPRERS
TH_1038|M.thermoresistible__bu TQ--------ERS