For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAVEPLYGTVIQFARLLWLIQGLKVSVRGIENLPASGGAVIAINHTSYLDFTFAGLPAYQQGLGRKVRFM AKQEVFDGKITGPIMRRLRHIPVDRQEGAASYEAAVRNLKDGELVGVYPEATISRSFEIKECKGGAARMS AEARVPIIPHIVWGSQRIWTKDCPKKLLRPKVPILVLVGEPIEPTLGVAQLKALLHFRMQHLLERAQELY GPHPAGEFWVPHRLGGGAPSLIKAARLDAEEATERAARRAQRAKVAE
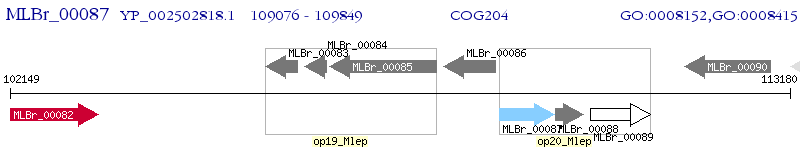
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00087 | - | - | 100% (257) | putative acyltransferase |
| M. leprae Br4923 | MLBr_00892 | - | 5e-14 | 32.85% (137) | putative acyltransferase |
| M. leprae Br4923 | MLBr_02427 | - | 1e-10 | 28.18% (181) | hypothetical protein MLBr_02427 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3846c | - | 1e-119 | 81.89% (254) | putative acyltransferase |
| M. gilvum PYR-GCK | Mflv_1153 | - | 1e-104 | 72.18% (248) | phospholipid/glycerol acyltransferase |
| M. tuberculosis H37Rv | Rv3816c | - | 1e-119 | 81.89% (254) | acyltransferase |
| M. abscessus ATCC 19977 | MAB_0165 | - | 1e-99 | 70.00% (250) | putative acyltransferase |
| M. marinum M | MMAR_5379 | - | 1e-114 | 78.35% (254) | acyltransferase |
| M. avium 104 | MAV_0202 | - | 1e-123 | 83.14% (255) | acyltransferase family protein |
| M. smegmatis MC2 155 | MSMEG_6409 | - | 1e-107 | 74.90% (251) | acyltransferase family protein |
| M. thermoresistible (build 8) | TH_0895 | - | 1e-104 | 73.60% (250) | POSSIBLE ACYLTRANSFERASE |
| M. ulcerans Agy99 | MUL_4999 | - | 1e-113 | 77.95% (254) | acyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5656 | - | 1e-107 | 75.30% (247) | phospholipid/glycerol acyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1153|M.gilvum_PYR-GCK --MEPVYGTAIQLARLVWRIQGLKFTVTGIENLPRTGGAVIAINHTSYFD
Mvan_5656|M.vanbaalenii_PYR-1 --MEPVYGTVIQLARLVWRVQGLKFTVTGVENLPTTGGAVIAINHTSYFD
TH_0895|M.thermoresistible__bu --VEPVYATVIQTARLVWRLQGLTFTVTGVENLPTTGGAVIAINHTSYFD
MSMEG_6409|M.smegmatis_MC2_155 --MEPVYGTVIQLARLAWRLQGLKFTVTGVENLPVTGGAVVAINHTSYFD
MLBr_00087|M.leprae_Br4923 MAVEPLYGTVIQFARLLWLIQGLKVSVRGIENLPASGGAVIAINHTSYLD
MAV_0202|M.avium_104 --MEPVYGTVIQLARLIWRVQGLKITITGVENLPKSGGAVIAINHTGYLD
MMAR_5379|M.marinum_M --MEPVYGTVIQLARLTWRIQGLRITVTGVENIPVQGGAVIAINHTGYLD
MUL_4999|M.ulcerans_Agy99 --MEPVYGTVIQLARLTWRIQGLRITVTGVENIPVQGGAVITINHTGYLD
Mb3846c|M.bovis_AF2122/97 --MEPVYGTVIRLARLSWRIQGLKITVTGVDNLPTSGGAVVAINHTSYLD
Rv3816c|M.tuberculosis_H37Rv --MEPVYGTVIRLARLSWRIQGLKITVTGVDNLPTSGGAVVAINHTSYLD
MAB_0165|M.abscessus_ATCC_1997 --MEPVYGTVIGLARTVWAIQGLKFTVTGVENLPTEGGAVVAINHTGYMD
:**:*.*.* ** * :*** .:: *::*:* ****::****.*:*
Mflv_1153|M.gilvum_PYR-GCK FTFAGLPAYRQHLGRKVRFMAKKEVFDNKISGPLMRGMKHIPVDRDSGAA
Mvan_5656|M.vanbaalenii_PYR-1 FTFAGLPAYQQHLGRKVRFMAKREVFDHKVTGPIMRSLRHIEVDRDSGAA
TH_0895|M.thermoresistible__bu FTFAGLPAYQQGLGRKVRFMAKKEVFDHKIGGPLMRSLRHIPVDRDSGAD
MSMEG_6409|M.smegmatis_MC2_155 FTFAGLPAYLQKQGRKVRFMAKKEVFDNKITGPIMRSLRHIEVDRHSGAP
MLBr_00087|M.leprae_Br4923 FTFAGLPAYQQGLGRKVRFMAKQEVFDGKITGPIMRRLRHIPVDRQEGAA
MAV_0202|M.avium_104 FTFAGLPAYKQGLGRKVRFMAKQEVFDHKITGPIMRSLRHISVNRQDGAA
MMAR_5379|M.marinum_M FTFGGLPAYQQRLGRKVRFMAKQEVFDQKITGPIMRKLRHIPVDRADGAA
MUL_4999|M.ulcerans_Agy99 FTFGGLPAYQQRLGRKVRFMAKQEVFDQKITGPIMRKLRHIPVDRADGAA
Mb3846c|M.bovis_AF2122/97 FTFAGLPAYQQGLGRKVRFMAKQEVFDHKITGPIMRSLRHIPVDRQDGSA
Rv3816c|M.tuberculosis_H37Rv FTFAGLPAYQQGLGRKVRFMAKQEVFDHKITGPIMRSLRHIPVDRQDGSA
MAB_0165|M.abscessus_ATCC_1997 FTFAGLPAFLQKKGRKVRFMAKKETFDNKITGPIMRGCRHISVDRVDGAA
***.****: * *********:*.** *: **:** :** *:* .*:
Mflv_1153|M.gilvum_PYR-GCK SFDEACRRLAAGEFVGVYPEATISRSFEIKEFKSGAARMAIAANVPIVPH
Mvan_5656|M.vanbaalenii_PYR-1 SFEEAVRKLKDGEFVGVYPEATISRSFEIKTFKSGAARMAIAAGVPIVPH
TH_0895|M.thermoresistible__bu SFAAACDALRAGELVGVYPEATISRSFEIKELKSGAARMAIECGVPIVPH
MSMEG_6409|M.smegmatis_MC2_155 SFEQACEYLKAGELVGVYPEATISRSFEIKELKSGAARMAIEADVPIIPH
MLBr_00087|M.leprae_Br4923 SYEAAVRNLKDGELVGVYPEATISRSFEIKECKGGAARMSAEARVPIIPH
MAV_0202|M.avium_104 SYEAAVRNLKDGELVGVYPEATISRSFEIKEFKSGAARMAVEAGVPIVPH
MMAR_5379|M.marinum_M SYDAAVRLLKAGELVGVYPEATISRSFEIKEFKSGAARMALEAGVPIVPH
MUL_4999|M.ulcerans_Agy99 SYDAAVRLLKAGELVGVYPEATISRSFEIKEFKSGAARMALEAGVPIVPH
Mb3846c|M.bovis_AF2122/97 SYDAAVRMLKAGELVGVYPEATISRSFEIKEFKTGAARMAIEAGVPIVPH
Rv3816c|M.tuberculosis_H37Rv SYDAAVRMLKAGELVGVYPEATISRSFEIKEFKTGAARMAIEAGVPIVPH
MAB_0165|M.abscessus_ATCC_1997 SYTEAVEKLKAGELVGVYPEATISRSFEIKEFKSGAARMAIEAGVPIVPH
*: * * **:**************** * *****: . ***:**
Mflv_1153|M.gilvum_PYR-GCK IVWGAQRIWTKGHPKNMKRPKVPIHIAVGTPIEPTLPAAELTTLLHSRMQ
Mvan_5656|M.vanbaalenii_PYR-1 IVWGAQRIWTKGHPKKMWRPKVPISIAVGKPIEPTLPAAELTTVLHSRMQ
TH_0895|M.thermoresistible__bu IVWGAQRIWTKGHPRKLLRPKVPISVAVGEPIQPTLPAPELTALLRSRMQ
MSMEG_6409|M.smegmatis_MC2_155 IVWGAQRIWTKGRPKNLYRPKVPIFIAVGEPIAPTLPAPELTALLHSRMQ
MLBr_00087|M.leprae_Br4923 IVWGSQRIWTKDCPKKLLRPKVPILVLVGEPIEPTLGVAQLKALLHFRMQ
MAV_0202|M.avium_104 IVWGAQRIWTKDHPKKLWRPKVPIIVLVGEPIQPTLGVEELKGLLHSRMQ
MMAR_5379|M.marinum_M IVWGAQRIWTKGHKKKLFRPKVPIVVVVGEPIEPTLPTAELSNLLHSRMQ
MUL_4999|M.ulcerans_Agy99 IVWGAQRIWTKGHKKKLFRPKVPIVVVVGEPIEPTLPTAELSNLLHSRMQ
Mb3846c|M.bovis_AF2122/97 IVWGAQRIWTKDRPKKLFRPKVPVTIVVGERIEPTLPTAELNGLLHSRMQ
Rv3816c|M.tuberculosis_H37Rv IVWGAQRIWTKDRPKKLFRPKVPVTIVVGERIEPTLPTAELNGLLHSRMQ
MAB_0165|M.abscessus_ATCC_1997 VIWGAQRIWTKGHPKHLGRTNTPISIAVGAPIAPTLPVSELTALLHARMQ
::**:******. ::: *.:.*: : ** * *** . :*. :*: ***
Mflv_1153|M.gilvum_PYR-GCK HLLAQVQDAYGPHPPGEFWVPHRMGGGAPTLAEANRMDAEEAAQKAARRS
Mvan_5656|M.vanbaalenii_PYR-1 HLLEQVQEDYGPHPPGEFWVPRRLGGGAPSLAEANQMDAEEAAAKAARRG
TH_0895|M.thermoresistible__bu HLLEQVQDSYGPHPPGEFWVPKRLGGGAPSLAEADQMDVEEARQKAARRA
MSMEG_6409|M.smegmatis_MC2_155 HLLEQVQDSYGPHPAGEFWVPHRLGGGAPSLAEANLMDAEEAAEKAARRA
MLBr_00087|M.leprae_Br4923 HLLERAQELYGPHPAGEFWVPHRLGGGAPSLIKAARLDAEEATERAARRA
MAV_0202|M.avium_104 HLLERAQEMYGPHPAGEFWVPHRLGGGAPSLAEAARLDAEEAAARAARRG
MMAR_5379|M.marinum_M HLLDQAQEQYGPHPSGEFWVPRRLGGSAPSLAEAAQMDAQEAAERAARRA
MUL_4999|M.ulcerans_Agy99 HLLDQAQEQYGPHPSGEFWVPRRLGGSAPSLAEAAQMDAQEAAERAARRA
Mb3846c|M.bovis_AF2122/97 HLLERAQELYGPHPAGEFWVPHRLGGGAPSLAEAARLDAQEAAVRAARRA
Rv3816c|M.tuberculosis_H37Rv HLLERAQELYGPHPAGEFWVPHRLGGGAPSLAEAARLDAQEAAVRAARRA
MAB_0165|M.abscessus_ATCC_1997 HLLLEVQDDYGPHPAGEYWVPHRLGGSAPTLAEAARWDAQDAAERKAQRA
*** ..*: *****.**:***:*:**.**:* :* *.::* : *:*.
Mflv_1153|M.gilvum_PYR-GCK ATPGIDTG-NAEAPD-----
Mvan_5656|M.vanbaalenii_PYR-1 GTPGLDAG-SAEAPG-----
TH_0895|M.thermoresistible__bu ARDAEGAG-TSESPG-----
MSMEG_6409|M.smegmatis_MC2_155 KQSGSGPAEPSGAPG-----
MLBr_00087|M.leprae_Br4923 QRAK-----VAE--------
MAV_0202|M.avium_104 QSAG-----APE--------
MMAR_5379|M.marinum_M QRAH-----PAGAPEQ----
MUL_4999|M.ulcerans_Agy99 QRAH-----PAGAPEQ----
Mb3846c|M.bovis_AF2122/97 QRAH-----PAGAPEQ----
Rv3816c|M.tuberculosis_H37Rv QRAH-----PAGAPEQ----
MAB_0165|M.abscessus_ATCC_1997 AREAEKQAQQAQKDGAQKES
.