For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSHTEGECPGADARSNSGTADAKSSSQSATKQPLPREVWVLVAANFVVALGYGVVAPVLPQYARHFGVS ISAATFVITAFAVMRLVGAPPAGMLVQRLGERRVYISGLIIVALSTAACAFAETYWQLLLFRSLGGIGSA MFTVSSLGLMIRISPPDARGRVAGLFSSGFMVGSVGGPILGALTAGFGLSAPFLIYGAALLVAATVVFVS LRHSTVVGQQDEDPEEPVPFRTVFRHRAYRAALFSNFATGWASFGLRIALVPLFIVEVLGQSPGAAGLAL TAFAVGNISVVIPSGHLSDRLGRRKLMIFGLTLAAASTALVGFTTSLPVFLVAAYIAGAATGIFVSPQQA AVADVVGNKSRGGTAVATFQMMADFGSIGGSLLVGLIAQYTSFGWAFLISGVILAVAAVGWIFAPETRPR ASTEHTQARPLGPEAGGEVP
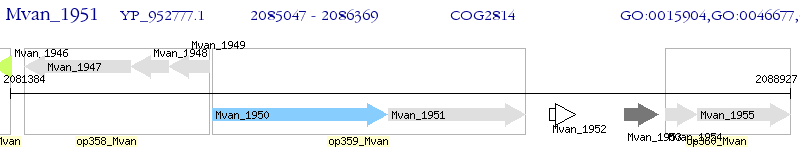
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1951 | - | - | 100% (440) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_1322 | - | 8e-19 | 24.62% (459) | major facilitator superfamily transporter |
| M. vanbaalenii PYR-1 | Mvan_5862 | - | 2e-17 | 26.13% (398) | major facilitator superfamily transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 1e-22 | 24.21% (413) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_4405 | - | 0.0 | 88.10% (420) | major facilitator transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 1e-22 | 24.21% (413) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_01562 | - | 4e-12 | 23.47% (409) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_3449c | - | 1e-125 | 52.40% (416) | putative transporter |
| M. marinum M | MMAR_0690 | - | 5e-32 | 29.07% (375) | putative transport protein |
| M. avium 104 | MAV_3974 | - | 1e-125 | 57.56% (377) | permease of the major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_2137 | - | 1e-173 | 73.44% (418) | permease of the major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_0476 | - | 1e-17 | 25.85% (383) | major facilitator superfamily protein MFS_1 |
| M. ulcerans Agy99 | MUL_3826 | - | 5e-17 | 23.06% (438) | hypothetical protein MUL_3826 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0805c|M.bovis_AF2122/97 -------------MLGNAMVEACPAEGDAPVPITPAGRPRSGQRS-YPDR
Rv0783c|M.tuberculosis_H37Rv -------------MLGNAMVEACPAEGDAPVPITPAGRPRSGQRS-YPDR
Mvan_1951|M.vanbaalenii_PYR-1 -------------------MTSH-TEGECPGADARSNSGTADAKSSSQSA
Mflv_4405|M.gilvum_PYR-GCK -------------------MSST-KEGDCP-------------------T
MSMEG_2137|M.smegmatis_MC2_155 -------------------MSSCPAEGDCS------------------AP
MAV_3974|M.avium_104 --------------------------------------------------
MAB_3449c|M.abscessus_ATCC_199 -------------------MSSC-REGNADN-----------------AT
TH_0476|M.thermoresistible__bu -------------------------------------------------V
MMAR_0690|M.marinum_M -------------------------------------------------M
MUL_3826|M.ulcerans_Agy99 ------------MMRPRATVAAATPTTDLAAG--------------RALI
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mb0805c|M.bovis_AF2122/97 LDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTG
Rv0783c|M.tuberculosis_H37Rv LDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQAVVAWTMTG
Mvan_1951|M.vanbaalenii_PYR-1 TKQPLPREVWVLVAANFVVALGYGVVAPVLPQYARHFGVSISAATFVITA
Mflv_4405|M.gilvum_PYR-GCK TKQRLPREVWVLVVANVVVALGYGVVAPVLPQYARHFGVSIAAATFVITA
MSMEG_2137|M.smegmatis_MC2_155 EKPKLPGEVWVLIVANAVIALGYGVVAPVLPQYARHFGVSISAATFVITA
MAV_3974|M.avium_104 -----------------MVALGYGVISPALPTFARSFGVSIKAVTFLVTV
MAB_3449c|M.abscessus_ATCC_199 VKPRLPWEIWVLVVASLVIALGFGVVAPALPQYARSFGVSVTAATAVVSS
TH_0476|M.thermoresistible__bu RRFAFP----LLAYAFAAVMVGTTVPTPMYAVYAERLGFGVLTTTVVFAG
MMAR_0690|M.marinum_M TDSSRRVVLAAVGFTFFIDMLVYGIVVPVLPRLAKPAGLEFLGVGVVFAC
MUL_3826|M.ulcerans_Agy99 TPRRRNFIFVALVLGILLSSLDQTIVAIALPTIVADLG-EAGRQSWVVTS
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
: : . .:
Mb0805c|M.bovis_AF2122/97 YMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVAPNILLLII
Rv0783c|M.tuberculosis_H37Rv YMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVAPNILLLII
Mvan_1951|M.vanbaalenii_PYR-1 FAVMRLVGAPPAGMLVQRLGERRVYISGLIIVALSTAACAFAETYWQLLL
Mflv_4405|M.gilvum_PYR-GCK FAVMRLVGAPPAGFLVQRLGERRVYTSGLIIVAVSTGACAFAETYWQLLL
MSMEG_2137|M.smegmatis_MC2_155 FALMRLCFAPATGMLIQRLGERRIYVNGLVIVALSTGACAFAQTYWQLLL
MAV_3974|M.avium_104 FSLSRLCFAPISGLLTERLGERRIYIGGLLIVAVSTAACAFSQAYWQLML
MAB_3449c|M.abscessus_ATCC_199 FAAFRLVFAPVAGALVQRLGERWVYMSGLLIVAASTGICAFVHSYWQLLV
TH_0476|M.thermoresistible__bu YTAGVLFALVVFGRWSDALGRRPMLLAGVGCAIVSALVFLIAGSVPQLIA
MMAR_0690|M.marinum_M YGLAYFVLTPVAGWLIDRRGSRSVFLSGAIILMLATLAFGYASSPVGLVT
MUL_3826|M.ulcerans_Agy99 YLLASTIATALVGKLGDMFGRKRVFQVAVLLFVAGSVSCGLTQSMTMLVA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLVI
: * : * : . .: *:
Mb0805c|M.bovis_AF2122/97 FRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPMLLGPVGGPI
Rv0783c|M.tuberculosis_H37Rv FRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPMLLGPVGGPI
Mvan_1951|M.vanbaalenii_PYR-1 FRSLGGIGSAMFTVSSLGLMIRISPP-DARGRVAGLFSSGFMVGSVGGPI
Mflv_4405|M.gilvum_PYR-GCK FRSLGGVGSAMFTVSALGLMIRISPA-DARGRVAGLFSSGFMIGSVGGPI
MSMEG_2137|M.smegmatis_MC2_155 FRSLGGIGSTMFFVSALGLMIRISPE-DARGRVAGMFSSAFLVGSVGGPV
MAV_3974|M.avium_104 CRVFSGVGSTMFYVSALGLMIHISPA-DARGRIAGLFTTSFMVGAVGGPA
MAB_3449c|M.abscessus_ATCC_199 FRSVGGVGSTMFTVSAAALLVRIAPE-EIRGRAQGLYGSSFLLGMVAGPA
TH_0476|M.thermoresistible__bu ARFVSGMSAGIFTGTATAAVIEAAPE-RRRSRAAAVATVANIGGLGAGPL
MMAR_0690|M.marinum_M CRVLQGGAAAATWVAGYATLVEIFPR-EQRGRAVGLASIGTALGALLGPA
MUL_3826|M.ulcerans_Agy99 SRALQGVGGGAITVTAIALIGEVVPL-RDRGRYQGILGAVIGIATIGGPL
MLBr_01562|M.leprae_Br4923 ARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLV
* . * .. . . : . .: . *
Mb0805c|M.bovis_AF2122/97 LGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASENFDYMGLL
Rv0783c|M.tuberculosis_H37Rv LGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASENFDYMGLL
Mvan_1951|M.vanbaalenii_PYR-1 LGALTAGFG----LSAPFLIYGAALLVAATVVFVSLR-------------
Mflv_4405|M.gilvum_PYR-GCK LGSLTAGFG----LAAPFLIYGAALLVAATVVFVSLR-------------
MSMEG_2137|M.smegmatis_MC2_155 LGSLTAGLG----LSAPFLIYGAALLVAAAVVFISLR-------------
MAV_3974|M.avium_104 VGGLAAGWG----LTAPFVVYGVAMLGVALVLFLGLR-------------
MAB_3449c|M.abscessus_ATCC_199 LGSAVVGLS----LSAPFIIYAAALVIVVMLVYFGLR-------------
TH_0476|M.thermoresistible__bu LAGILVQYA-------PWPLHLTFWVHIVLMVLAAAA-------------
MMAR_0690|M.marinum_M LGGLAYEASG---PHAPFLGAAGLSALAVAALAASLP-------------
MUL_3826|M.ulcerans_Agy99 LGGYFTDCLGWRWAFWINLPVSAVVICVATAAIPALAATTRPVIDYAGIM
MLBr_01562|M.leprae_Br4923 VGGALTEVS----WRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAG
:..
Mb0805c|M.bovis_AF2122/97 LLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFVAHSWYRTE
Rv0783c|M.tuberculosis_H37Rv LLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFVAHSWYRTE
Mvan_1951|M.vanbaalenii_PYR-1 ------HSTVVGQQDEDPEEPVPFRTVFRHRAYRAALFSNFATG------
Mflv_4405|M.gilvum_PYR-GCK ------NSAVVGQADDDVQVPIPFRTVFRHRAYKAALFSNFATG------
MSMEG_2137|M.smegmatis_MC2_155 ------HSSLAAPAPDDGPK-VTVRTALRNGAYRAALVSNLATG------
MAV_3974|M.avium_104 ------NSALAAPPPP-TRSTVTMREALRVRAYQSALLSNFATG------
MAB_3449c|M.abscessus_ATCC_199 ------RSTLLDVADEHHGTPVRLTSALRHRTFLAALMSNFSAG------
TH_0476|M.thermoresistible__bu -------IAVAPETSPRTGRLGVQRLSLPPQVRKVFAIASLAAFAG----
MMAR_0690|M.marinum_M ----------QAAGGQASGPGMWSGERAFPGLAVPWLVAGLAVG------
MUL_3826|M.ulcerans_Agy99 FIG-LSLAALTLATSLGGSVYAWGSAPIIGLFTAAGVTLAVFVWVETIAA
MLBr_01562|M.leprae_Br4923 AILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAE
.
Mb0805c|M.bovis_AF2122/97 HPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQS-PMQ
Rv0783c|M.tuberculosis_H37Rv HPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQVLHQS-PMQ
Mvan_1951|M.vanbaalenii_PYR-1 ----------------------WASFGLR--IALVPLFIVEVLGQS-PGA
Mflv_4405|M.gilvum_PYR-GCK ----------------------WASFGLR--IALVPLFVVEVLGEG-PGA
MSMEG_2137|M.smegmatis_MC2_155 ----------------------WSAFGLR--IALVPLFIAEVFHRG-PGI
MAV_3974|M.avium_104 ----------------------WSAFGLR--MALVPLFVSDVMGRG-IGT
MAB_3449c|M.abscessus_ATCC_199 ----------------------WAIFGIR--GALLPLLVVEALHQR-PGA
TH_0476|M.thermoresistible__bu ----------------------FAVTGLF--TAVAPSFLAEVIGVRNHAV
MMAR_0690|M.marinum_M -------------------------LALGAVEATLPIDLVARFDAS-GVT
MUL_3826|M.ulcerans_Agy99 QPILPIRLFAAPVFSVCCVLAFVVGFAMLGALIFVPTFMQYVNGVS-ATA
MLBr_01562|M.leprae_Br4923 NPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYS-ALR
: :
Mb0805c|M.bovis_AF2122/97 SGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIV-LVGIMLIAAGLGTFAFG
Rv0783c|M.tuberculosis_H37Rv SGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIV-LVGIMLIAAGLGTFAFG
Mvan_1951|M.vanbaalenii_PYR-1 AGLALTAFAVGNISVVIPSGHLSDRLGRRKLM-IFGLTLAAASTALVGFT
Mflv_4405|M.gilvum_PYR-GCK AGLALTAFAVGNIAVVIPSGHLSDRLGRRKLL-IFGLMLAAVSTALVGFT
MSMEG_2137|M.smegmatis_MC2_155 AGLALATFAIGNVGAVIPSGYLSDRVGRRVLL-IIGLTAAGVATAVVGLT
MAV_3974|M.avium_104 IGVVLAAFAGGNALAVVPSGYLSDRMGRRTLL-IVGLVTSGAATVWLGFV
MAB_3449c|M.abscessus_ATCC_199 AGLVFAAFAAGDVSTAFLAGSWSDDIGRKPLV-VAGLVVCGSTVVAMGFV
TH_0476|M.thermoresistible__bu AGAVAASIFAASAVTQVLGASVSPR--RAVAV-GCALLIVGMVFLAAALV
MMAR_0690|M.marinum_M VGLLFALTTVAFIVTSQYAGRLSDAGRRAPVL-AAGWVTLGVALAVMGVP
MUL_3826|M.ulcerans_Agy99 SGLRILPMVIGMLITSIGSGSMVGRTGRYKIFPVLGTALMTLAFLLMSRT
MLBr_01562|M.leprae_Br4923 AGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFM
* . .. . .
Mb0805c|M.bovis_AF2122/97 VARQADYLPILPTGLAIMGMGMGCSMM---PLSGAAVQTLAPHQIARGST
Rv0783c|M.tuberculosis_H37Rv VARQADYLPILPTGLAIMGMGMGCSMM---PLSGAAVQTLAPHQIARGST
Mvan_1951|M.vanbaalenii_PYR-1 TS-----LPVFLVAAYIAGAATGIFVS---PQQAAVADVVGNKSRGGTAV
Mflv_4405|M.gilvum_PYR-GCK TS-----LPVFLVAAVIAGAATGIFVS---PQQAAVADVVGNKSRGGPAV
MSMEG_2137|M.smegmatis_MC2_155 DG-----LTPFLVSALVAGFATGIFTA---PQQAAVADIIGNKARGGTAV
MAV_3974|M.avium_104 AS-----LPVFLVAAGVVGVVTGIYMS---PLQAAVADILGNEARAGLPV
MAB_3449c|M.abscessus_ATCC_199 SS-----LPVLVVLSLIGGIGAGLYAS---PQQAAVADVVGSRGRAGTAL
TH_0476|M.thermoresistible__bu WSS----LPALIATAVVAGAGQGLSFS---RGLAAVSARTPSDQR-GEVS
MMAR_0690|M.marinum_M SA-----VGIQAAALVLLGAGLGAMIATIMPALADAMESAGGAQRPGTAA
MUL_3826|M.ulcerans_Agy99 DESTS--AAVQSVYLLILGSAIGMSSQ---VLVIIVQNTSEFEDLGVATS
MLBr_01562|M.leprae_Br4923 NRGVP-YFPNLLAPIVVGGIGIGMAVV---PLTLSAIAGVGFDRIGPVSA
. : * * .
Mb0805c|M.bovis_AF2122/97 LISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAA
Rv0783c|M.tuberculosis_H37Rv LISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGRGAA
Mvan_1951|M.vanbaalenii_PYR-1 ATFQMMADFGSIGGSLLVGLIAQYTSFGWAFLISGVILAVAAVGWIFAPE
Mflv_4405|M.gilvum_PYR-GCK ATFQMMADFGSIGGSLLVGLIAQYTSYGWAFLVSGIILAVAAIGWFFAPE
MSMEG_2137|M.smegmatis_MC2_155 ATFQMMADVGSIGGSLLVGLIAQYLSFAWAFGISGGMLLLAALCWTFAPE
MAV_3974|M.avium_104 ATVQMMSDLGAIVGSMAVGWAAEQIGYGWGFFISGVVLLIAAVGWVMAPE
MAB_3449c|M.abscessus_ATCC_199 ATFQMTSDIGLVVGPVVAGAIAEHTSYGWAFVVGGAMPLIAAVAWVFAPE
TH_0476|M.thermoresistible__bu STYFVVAYVALLVPVIGEGWAAHHWGLRTAGISFAVGVAVLAIGCLIAIL
MMAR_0690|M.marinum_M AAYNLAFSAGTVAGGPLAGWMADLRSFQDAAMLIAVVTLTCGVLAALVHA
MUL_3826|M.ulcerans_Agy99 GVSLFRTIGGSFGAAIFGSLFVNFLNSRLASALTATEVASSPEALHRQSP
MLBr_01562|M.leprae_Br4923 IALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGY
. . .
Mb0805c|M.bovis_AF2122/97 VDPSSLPRQTNFAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQQA
Rv0783c|M.tuberculosis_H37Rv VDPSSLPRQTNFAAQLLHDLSHAYAVVFVIATALVVSTLIPAAFLPKQQA
Mvan_1951|M.vanbaalenii_PYR-1 TR-PRASTEHTQARPLGPEAGGEVP-------------------------
Mflv_4405|M.gilvum_PYR-GCK TR-PTPSTEHTPGRPLGPEAGGEVP-------------------------
MSMEG_2137|M.smegmatis_MC2_155 TR-PRPSAEHTPARPLGPEAGGEVP-------------------------
MAV_3974|M.avium_104 TR-TATELEADLMAASRTSSRCEQQL------------------------
MAB_3449c|M.abscessus_ATCC_199 TLGRLASPQVRSMQEVAEDVGGPER-------------------------
TH_0476|M.thermoresistible__bu IQESRDPQAI----------------------------------------
MMAR_0690|M.marinum_M AVVRRAVSDA----------------------------------------
MUL_3826|M.ulcerans_Agy99 AVAAPIVHAYAGSLAQVFLCAAPVAAIGFVLALFMREVPLRDIHNRDHDA
MLBr_01562|M.leprae_Br4923 TYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL-------
Mb0805c|M.bovis_AF2122/97 SHRRAPLLSA---------------------------------
Rv0783c|M.tuberculosis_H37Rv SHRRAPLLSA---------------------------------
Mvan_1951|M.vanbaalenii_PYR-1 -------------------------------------------
Mflv_4405|M.gilvum_PYR-GCK -------------------------------------------
MSMEG_2137|M.smegmatis_MC2_155 -------------------------------------------
MAV_3974|M.avium_104 -------------------------------------------
MAB_3449c|M.abscessus_ATCC_199 -------------------------------------------
TH_0476|M.thermoresistible__bu -------------------------------------------
MMAR_0690|M.marinum_M -------------------------------------------
MUL_3826|M.ulcerans_Agy99 RVPSGDGSGQQNKLEGDLDTGQSVGAPGKEPDFGVDPAMGPVR
MLBr_01562|M.leprae_Br4923 -------------------------------------------