For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAVDRVRRQDVIPERPAPETPRAHVCVLASLNYPDISAEDIALIRRFTRVALNTLIELGASYELWDTSA TLENPSAAADFDGLLLLGGGDIDPTCYGGGARHPKSYGVDARADRDAFAVIAAAEAAGRPIFGICRGSQL LNVARGGTIIGDIVDYALHHGAPGEPEFVDEPIDIMPGTRLASILGADRVIGRSGHHQAVDTLGRGLVVA ARALDGITEGVEDPDKFYLGVQWHPEDDDGPEADRMRLFGAFVTAAERSRENTAAVASQG
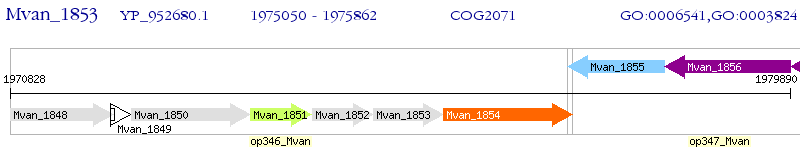
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1853 | - | - | 100% (270) | peptidase C26 |
| M. vanbaalenii PYR-1 | Mvan_2279 | - | 2e-31 | 42.47% (186) | peptidase C26 |
| M. vanbaalenii PYR-1 | Mvan_1852 | - | 4e-29 | 40.10% (192) | peptidase C26 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2884c | - | 3e-28 | 42.94% (177) | amidotransferase |
| M. gilvum PYR-GCK | Mflv_4064 | - | 2e-31 | 43.32% (187) | peptidase C26 |
| M. tuberculosis H37Rv | Rv2859c | - | 3e-28 | 42.94% (177) | amidotransferase |
| M. leprae Br4923 | MLBr_01573 | - | 4e-31 | 44.26% (183) | putative amidotransferase |
| M. abscessus ATCC 19977 | MAB_0032 | - | 2e-09 | 28.49% (186) | para-aminobenzoate synthase component II |
| M. marinum M | MMAR_1844 | - | 1e-30 | 42.54% (181) | amidotransferase |
| M. avium 104 | MAV_3717 | - | 1e-28 | 46.63% (178) | class I glutamine amidotransferase, putative |
| M. smegmatis MC2 155 | MSMEG_6690 | - | 1e-118 | 77.57% (263) | glutamine amidotransferase class-I |
| M. thermoresistible (build 8) | TH_0292 | pabA | 8e-07 | 28.34% (187) | POSSIBLE ANTHRANILATE SYNTHASE COMPONENT II TRPG (GLUTAMINE |
| M. ulcerans Agy99 | MUL_2093 | - | 3e-30 | 42.54% (181) | amidotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1853|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6690|M.smegmatis_MC2_155 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
Rv2859c|M.tuberculosis_H37Rv MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0292|M.thermoresistible__bu --------------------------------------------------
Mvan_1853|M.vanbaalenii_PYR-1 -----MTAVDRVRRQDVIPERPAPETPRAHVCVLASLNYPDISAEDIALI
MSMEG_6690|M.smegmatis_MC2_155 -------MTIRVRREDVIPNPSEPTSVRAHVCVVASLNFPGINEHDAGLV
Mb2884c|M.bovis_AF2122/97 RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADYFEGITMA
Rv2859c|M.tuberculosis_H37Rv RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADYFEGITMA
MAV_3717|M.avium_104 -----------------MGLTAYLERVQTGIWDIPAGYLPADYFEGVTRA
MLBr_01573|M.leprae_Br4923 --------MDLSGSRPVVGLTAYLEQVHTGLWDVPAGYLPADYFQGVAMA
MMAR_1844|M.marinum_M ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASYFDGVTMA
MUL_2093|M.ulcerans_Agy99 ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASYFDGVTMA
Mflv_4064|M.gilvum_PYR-GCK --------MSVS-SRPVIGLTTYLQQAQTGVWDVRASFLPAIYFEGVGMA
MAB_0032|M.abscessus_ATCC_1997 ------------------------------MRILVVDNYDSFVFNLVQYL
TH_0292|M.thermoresistible__bu ------------------------------VQILVVDNYDSFVFNLVQYL
: : .
Mvan_1853|M.vanbaalenii_PYR-1 RRFTRVALNTLIELGASYELWDTSATLENPSAAADFDGLLLLGGGDIDPT
MSMEG_6690|M.smegmatis_MC2_155 RRFTRTALATLVSLGASFELWDTTQPLENPEAAADFDGLLLLGGGDVDGS
Mb2884c|M.bovis_AF2122/97 GGVAVLLPPQPVDPESVGCVLDS------------LHALVITGGYDLDPA
Rv2859c|M.tuberculosis_H37Rv GGVAVLLPPQPVDPESVGCVLDS------------LHALVITGGYDLDPA
MAV_3717|M.avium_104 GGIAVLLPPQPVDTGIVGSLLDG------------LHALVITGGYDLDPA
MLBr_01573|M.leprae_Br4923 GGIAVLLPPQPVDPEIAGLALDG------------LDGLVITGGYDVDPA
MMAR_1844|M.marinum_M GGIAVLLPPQRADADAANRVLDS------------LDALVITGGKDVDPA
MUL_2093|M.ulcerans_Agy99 GGIAVLLPPQRADADAANRVLDS------------LDALVITGGKDVDPA
Mflv_4064|M.gilvum_PYR-GCK GGIASLLPPQPVDDTIAERVLDG------------IDGLIITGGRDVDPS
MAB_0032|M.abscessus_ATCC_1997 GQLGVDATVWRNDDPRLADT-----------------DAVA---ADFDGV
TH_0292|M.thermoresistible__bu GQLGVDAQVWRNDDARLGTE-----------------AAIADAVAAVDGV
. . : .*
Mvan_1853|M.vanbaalenii_PYR-1 CYGGGARHPKSYGVDAR---ADRDAFAVIAAAEAAGRPIFGICRGSQLLN
MSMEG_6690|M.smegmatis_MC2_155 CYGSCEPIPNSYGVDLR---ADRDAFAVIAAAEAAGRPIFGICRGSQLLN
Mb2884c|M.bovis_AF2122/97 AYG-QEPHPATDHPRPG---RDAWEFALLRGALQRGMPVLGICRGTQVLN
Rv2859c|M.tuberculosis_H37Rv AYG-QEPHPATDHPRPG---RDAWEFALLRGALQRGMPVLGICRGTQVLN
MAV_3717|M.avium_104 GYG-QRPHPSTDAPRTD---RDAWEFALLRGALQRGLPVLGICRGAQVLN
MLBr_01573|M.leprae_Br4923 TYG-QRPHPSTDEPRTT---RDSWEFALFEAALQRGLPVLGICRGAQLLN
MMAR_1844|M.marinum_M AYG-QQAHPATDEPAPI---RDSWEFALLRGALQRGLPVLGICRGAQVLN
MUL_2093|M.ulcerans_Agy99 AYG-QQAHPATDEPAPI---RDSWEFALLRGALQRGLPVLGICRGAQVLN
Mflv_4064|M.gilvum_PYR-GCK TYG-AQRHPRTDEPDTDSRARDAFEFALVRAALRRAMPVLGICRGAQVLN
MAB_0032|M.abscessus_ATCC_1997 LIS---PGPGTPESAGA-------SIAMVHSCAKTGTPLLGVCLGHQAIG
TH_0292|M.thermoresistible__bu LLS---PGPGTPERAGA-------SIPVVRACAAAGTPLLGVCLGHQAIG
. * : :.:. .. . *::*:* * * :.
Mvan_1853|M.vanbaalenii_PYR-1 VARGGTIIGDIVDYALHHGA-PGEPE-FVDEPIDIMPGTRLASILGADRV
MSMEG_6690|M.smegmatis_MC2_155 VARGGSLIGDIVDYKLHHGG-PGEPV-FVDEPIDVVPGTRLRSILGTDRV
Mb2884c|M.bovis_AF2122/97 VALGGTLHQHLPDILGHSGHRAGNGV-FTRLPVHTASGTRLAELIG-ESA
Rv2859c|M.tuberculosis_H37Rv VALGGTLHQHLPDILGHSGHRAGNGV-FTRLPVHTASGTRLAELIG-ESA
MAV_3717|M.avium_104 VAFGGTLHQHLPDVLGHGGHRAGNGV-FSTLPVRTVAGTRLAALLG-ETV
MLBr_01573|M.leprae_Br4923 IALGGTLHQHLPEVIGHSKHWVGNAV-FNNLLVRTVPGTRLAAVLG-EFV
MMAR_1844|M.marinum_M VAFGGTLHQHLPDVLGHNGHRAGNAV-FSSLPVRTVPGTRLATLIG-ESA
MUL_2093|M.ulcerans_Agy99 VAFGGTPHQHLPDVLGHNGHRAGNAV-FSSLPVRTVPGTRLATLIG-ESA
Mflv_4064|M.gilvum_PYR-GCK VALGGTLHQHVPDVVGHTRHQQGNAV-FTTSSIATVPGSRVAALVG-PDT
MAB_0032|M.abscessus_ATCC_1997 VAFGGTVDRAPELLHGKTSTVEHEDAGVLRGLPNPFTATRYHSLTILPDT
TH_0292|M.thermoresistible__bu VAFGATVDRAPELLHGKTSTVHHTDVGVLQGLPDPFTATRYHSLTILPQT
:* *.: : . ..:* : .
Mvan_1853|M.vanbaalenii_PYR-1 IGRSGHHQAVDTLGRGLVVAARALDGITEGVEDPD-------KFYLGVQW
MSMEG_6690|M.smegmatis_MC2_155 IGRNGHHQAVDRLGRGLVVAARALDGITEGVEDPD-------RFYLGVQW
Mb2884c|M.bovis_AF2122/97 DVPCYHHQAIDQVGEGLVVSAVDVDGVIEALELPG------DTFVLAVQW
Rv2859c|M.tuberculosis_H37Rv DVPCYHHQAIDQVGEGLVVSAVDVDGVIEALELPG------DTFVLAVQW
MAV_3717|M.avium_104 DAPCYHHQAIDKLGDGLVVSAWDPDGVVEAVELPG------DAFVLAVQW
MLBr_01573|M.leprae_Br4923 EARCYHHQSIDKLGDGLVVSAWDADGVVEAVELPG------DAFVLGVQW
MMAR_1844|M.marinum_M QVRCYHHQAVAEVGEGLVVSAWDVDGVVEALELPG------ENFVLAVQW
MUL_2093|M.ulcerans_Agy99 QVRCYHHQAVAEVGEGLVVSAFDVDGVVEALELPG------ENFVLAVQW
Mflv_4064|M.gilvum_PYR-GCK EAQCYHHQAIDRLGDGLVVSASDTDGVIEAVEVDLS--AHPDHWVIAVQW
MAB_0032|M.abscessus_ATCC_1997 VP--EGLRVIARTASGVIMGVMHTEHPIHGVQFHPESILTEGGHRMLANW
TH_0292|M.thermoresistible__bu LP--AELEVTARTAGGVIMGVRHRELPIHGVQFHPESILTEGGHRMLANW
. . *:::.. : ..:: : .:*
Mvan_1853|M.vanbaalenii_PYR-1 HPEDDDGPEADRMRLFGAFVTAAERSRENTAAVASQG-
MSMEG_6690|M.smegmatis_MC2_155 HPEDTDGPEADRMRLFEAFVGAAEQARQSRALAAAPAE
Mb2884c|M.bovis_AF2122/97 HPEKS----LDDLRLFKALVDAASGYAGRQSQAEPR--
Rv2859c|M.tuberculosis_H37Rv HPEKS----LDDLRLFKALVDAASGYAGRQSQAEPR--
MAV_3717|M.avium_104 HPEQS----LHDLRLFTAIVDAARSYAGVAR-------
MLBr_01573|M.leprae_Br4923 HPEKA----LSDLRLFTAIVDAACRYANSRCVEYS---
MMAR_1844|M.marinum_M HPEES----LEDLRLFSAVVDAARSRRSG---------
MUL_2093|M.ulcerans_Agy99 HPEES----LEDLRLFSAVVDAARSRRSG---------
Mflv_4064|M.gilvum_PYR-GCK HPEER----LDDLRLFAGLVGAAGVYATQKTQKVNR--
MAB_0032|M.abscessus_ATCC_1997 LTYCGAPLDEPLVRSLEAEVAAALNGVTGRVAALGG--
TH_0292|M.thermoresistible__bu LGFCGAAPDDALVRRLEDEVAETVRAATTRSSA-----
:* : * :