For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TIRVRREDVIPNPSEPTSVRAHVCVVASLNFPGINEHDAGLVRRFTRTALATLVSLGASFELWDTTQPLE NPEAAADFDGLLLLGGGDVDGSCYGSCEPIPNSYGVDLRADRDAFAVIAAAEAAGRPIFGICRGSQLLNV ARGGSLIGDIVDYKLHHGGPGEPVFVDEPIDVVPGTRLRSILGTDRVIGRNGHHQAVDRLGRGLVVAARA LDGITEGVEDPDRFYLGVQWHPEDTDGPEADRMRLFEAFVGAAEQARQSRALAAAPAE*
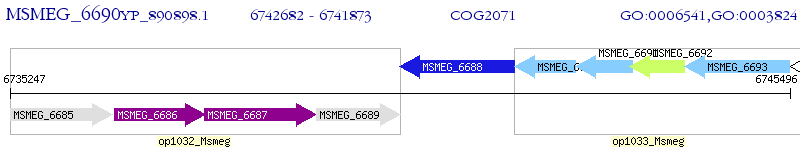
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6690 | - | - | 100% (269) | glutamine amidotransferase class-I |
| M. smegmatis MC2 155 | MSMEG_2596 | - | 2e-32 | 45.76% (177) | peptidase C26 |
| M. smegmatis MC2 155 | MSMEG_6691 | - | 4e-28 | 39.79% (191) | glutamine amidotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2884c | - | 7e-29 | 40.20% (204) | amidotransferase |
| M. gilvum PYR-GCK | Mflv_4064 | - | 3e-31 | 44.62% (186) | peptidase C26 |
| M. tuberculosis H37Rv | Rv2859c | - | 7e-29 | 40.20% (204) | amidotransferase |
| M. leprae Br4923 | MLBr_01573 | - | 2e-31 | 42.65% (204) | putative amidotransferase |
| M. abscessus ATCC 19977 | MAB_0032 | - | 5e-07 | 26.60% (188) | para-aminobenzoate synthase component II |
| M. marinum M | MMAR_1844 | - | 5e-29 | 41.99% (181) | amidotransferase |
| M. avium 104 | MAV_3717 | - | 3e-29 | 44.62% (195) | class I glutamine amidotransferase, putative |
| M. thermoresistible (build 8) | TH_0817 | carA | 6e-07 | 30.67% (163) | PROBABLE CARBAMOYL-PHOSPHATE SYNTHASE SMALL CHAIN CARA |
| M. ulcerans Agy99 | MUL_2093 | - | 2e-28 | 41.44% (181) | amidotransferase |
| M. vanbaalenii PYR-1 | Mvan_1853 | - | 1e-118 | 77.57% (263) | peptidase C26 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6690|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1853|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 --------------------------------------------------
Rv2859c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0817|M.thermoresistible__bu MTMTASTQATTAATRAVLVLEDGRVFTGVAFGAVGETLGEAVFTTGMSGY
MSMEG_6690|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_1853|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 -----------------------MDLSASRSDGGDPLRPASPRLRSPVSD
Rv2859c|M.tuberculosis_H37Rv -----------------------MDLSASRSDGGDPLRPASPRLRSPVSD
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0817|M.thermoresistible__bu QEALTDPSFHRQIVIATAPHIGNTGWNDEDSESRGDKIWVAGYAVRDPSP
MSMEG_6690|M.smegmatis_MC2_155 ------------------------------MTIRVRREDVIPNPSEPTSV
Mvan_1853|M.vanbaalenii_PYR-1 ----------------------------MTAVDRVRRQDVIPERPAPETP
Mb2884c|M.bovis_AF2122/97 GGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPLGASRPVVGLTAYLEQV
Rv2859c|M.tuberculosis_H37Rv GGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPLGASRPVVGLTAYLEQV
MAV_3717|M.avium_104 ----------------------------------------MGLTAYLERV
MLBr_01573|M.leprae_Br4923 -------------------------------MDLSGSRPVVGLTAYLEQV
MMAR_1844|M.marinum_M -----------------------------MVLNAAGPGPIVGMTTYLDQA
MUL_2093|M.ulcerans_Agy99 -----------------------------MVLNAAGPGPIVGMTTYLDQA
Mflv_4064|M.gilvum_PYR-GCK -------------------------------MSVS-SRPVIGLTTYLQQA
MAB_0032|M.abscessus_ATCC_1997 -----------------------------------------MRILVVDNY
TH_0817|M.thermoresistible__bu RASNWRATRTLEEELVRQNIVGIAGIDTRAVVRHLRTRGTMKAGVFSGPA
MSMEG_6690|M.smegmatis_MC2_155 RAHVCVVASLNFPGINEHDAGLVRRFTRTALATLVSLGASFELWDTTQP-
Mvan_1853|M.vanbaalenii_PYR-1 RAHVCVLASLNYPDISAEDIALIRRFTRVALNTLIELGASYELWDTSAT-
Mb2884c|M.bovis_AF2122/97 RTGVWDIPAGYLPADYFEGITMAGGVAVLLPPQPVDPESVGCVLDS----
Rv2859c|M.tuberculosis_H37Rv RTGVWDIPAGYLPADYFEGITMAGGVAVLLPPQPVDPESVGCVLDS----
MAV_3717|M.avium_104 QTGIWDIPAGYLPADYFEGVTRAGGIAVLLPPQPVDTGIVGSLLDG----
MLBr_01573|M.leprae_Br4923 HTGLWDVPAGYLPADYFQGVAMAGGIAVLLPPQPVDPEIAGLALDG----
MMAR_1844|M.marinum_M KTGVWDVSASFLPASYFDGVTMAGGIAVLLPPQRADADAANRVLDS----
MUL_2093|M.ulcerans_Agy99 KTGVWDVSASFLPASYFDGVTMAGGIAVLLPPQRADADAANRVLDS----
Mflv_4064|M.gilvum_PYR-GCK QTGVWDVRASFLPAIYFEGVGMAGGIASLLPPQPVDDTIAERVLDG----
MAB_0032|M.abscessus_ATCC_1997 DSFVFNLVQ------YLGQLGVDATVWRNDDPRLADTDAVAADFDG----
TH_0817|M.thermoresistible__bu LADIDELVTRVREQPSMLGANLSGEVTTPEPYVLEPEGPHRFVIAAIDLG
: : : .
MSMEG_6690|M.smegmatis_MC2_155 ----LENPEAAADFDGLLLLGGGDVDGSCYGSCEPIPNSYGVDLR---AD
Mvan_1853|M.vanbaalenii_PYR-1 ----LENPSAAADFDGLLLLGGGDIDPTCYGGGARHPKSYGVDAR---AD
Mb2884c|M.bovis_AF2122/97 -------------LHALVITGGYDLDPAAYG-QEPHPATDHPRPG---RD
Rv2859c|M.tuberculosis_H37Rv -------------LHALVITGGYDLDPAAYG-QEPHPATDHPRPG---RD
MAV_3717|M.avium_104 -------------LHALVITGGYDLDPAGYG-QRPHPSTDAPRTD---RD
MLBr_01573|M.leprae_Br4923 -------------LDGLVITGGYDVDPATYG-QRPHPSTDEPRTT---RD
MMAR_1844|M.marinum_M -------------LDALVITGGKDVDPAAYG-QQAHPATDEPAPI---RD
MUL_2093|M.ulcerans_Agy99 -------------LDALVITGGKDVDPAAYG-QQAHPATDEPAPI---RD
Mflv_4064|M.gilvum_PYR-GCK -------------IDGLIITGGRDVDPSTYG-AQRHPRTDEPDTDSRARD
MAB_0032|M.abscessus_ATCC_1997 -----------------VLISPGPGTPESAG-------------------
TH_0817|M.thermoresistible__bu IKTNTPRNFVQRGVRTHVMPAGTTFDELRDLKPDGVFLSNGPGDP--ATA
:: .
MSMEG_6690|M.smegmatis_MC2_155 RDAFAVIAAAEAAGRPIFGICRGSQLLNVARGGSLIGDIVDYKLHHGG-P
Mvan_1853|M.vanbaalenii_PYR-1 RDAFAVIAAAEAAGRPIFGICRGSQLLNVARGGTIIGDIVDYALHHGA-P
Mb2884c|M.bovis_AF2122/97 AWEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPDILGHSGHRA
Rv2859c|M.tuberculosis_H37Rv AWEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPDILGHSGHRA
MAV_3717|M.avium_104 AWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPDVLGHGGHRA
MLBr_01573|M.leprae_Br4923 SWEFALFEAALQRGLPVLGICRGAQLLNIALGGTLHQHLPEVIGHSKHWV
MMAR_1844|M.marinum_M SWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPDVLGHNGHRA
MUL_2093|M.ulcerans_Agy99 SWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTPHQHLPDVLGHNGHRA
Mflv_4064|M.gilvum_PYR-GCK AFEFALVRAALRRAMPVLGICRGAQVLNVALGGTLHQHVPDVVGHTRHQQ
MAB_0032|M.abscessus_ATCC_1997 -ASIAMVHSCAKTGTPLLGVCLGHQAIGVAFGGTVDRAPELLHGKTSTVE
TH_0817|M.thermoresistible__bu DHVVAVTRSVLEAGIPLFGICFGNQILGRALGRSTYKMVFGHRGHNVPVI
.*: . . *::*:* * * :. * * : :
MSMEG_6690|M.smegmatis_MC2_155 GEPV-FVDEPIDVVPGTRLRSILGTDRVIGRNGHHQAVDRLGRGLVVAAR
Mvan_1853|M.vanbaalenii_PYR-1 GEPE-FVDEPIDIMPGTRLASILGADRVIGRSGHHQAVDTLGRGLVVAAR
Mb2884c|M.bovis_AF2122/97 GNGV-FTRLPVHTASGTRLAELIGES-ADVPCYHHQAIDQVGEGLVVSAV
Rv2859c|M.tuberculosis_H37Rv GNGV-FTRLPVHTASGTRLAELIGES-ADVPCYHHQAIDQVGEGLVVSAV
MAV_3717|M.avium_104 GNGV-FSTLPVRTVAGTRLAALLGET-VDAPCYHHQAIDKLGDGLVVSAW
MLBr_01573|M.leprae_Br4923 GNAV-FNNLLVRTVPGTRLAAVLGEF-VEARCYHHQSIDKLGDGLVVSAW
MMAR_1844|M.marinum_M GNAV-FSSLPVRTVPGTRLATLIGES-AQVRCYHHQAVAEVGEGLVVSAW
MUL_2093|M.ulcerans_Agy99 GNAV-FSSLPVRTVPGTRLATLIGES-AQVRCYHHQAVAEVGEGLVVSAF
Mflv_4064|M.gilvum_PYR-GCK GNAV-FTTSSIATVPGSRVAALVGPD-TEAQCYHHQAIDRLGDGLVVSAS
MAB_0032|M.abscessus_ATCC_1997 HEDAGVLRGLPNPFTATRYHSLTILP--DTVPEGLRVIARTASGVIMGVM
TH_0817|M.thermoresistible__bu DHRT----GRVAITAQNHGFALEGEA-------GEVFDTPFGRAEISHTC
. . .: : . . : .
MSMEG_6690|M.smegmatis_MC2_155 ALDGITEGVEDPD-------RFYLGVQWHPEDTDGPEADRMRLFEAFVGA
Mvan_1853|M.vanbaalenii_PYR-1 ALDGITEGVEDPD-------KFYLGVQWHPEDDDGPEADRMRLFGAFVTA
Mb2884c|M.bovis_AF2122/97 DVDGVIEALELPG------DTFVLAVQWHPEKS----LDDLRLFKALVDA
Rv2859c|M.tuberculosis_H37Rv DVDGVIEALELPG------DTFVLAVQWHPEKS----LDDLRLFKALVDA
MAV_3717|M.avium_104 DPDGVVEAVELPG------DAFVLAVQWHPEQS----LHDLRLFTAIVDA
MLBr_01573|M.leprae_Br4923 DADGVVEAVELPG------DAFVLGVQWHPEKA----LSDLRLFTAIVDA
MMAR_1844|M.marinum_M DVDGVVEALELPG------ENFVLAVQWHPEES----LEDLRLFSAVVDA
MUL_2093|M.ulcerans_Agy99 DVDGVVEALELPG------ENFVLAVQWHPEES----LEDLRLFSAVVDA
Mflv_4064|M.gilvum_PYR-GCK DTDGVIEAVEVDLS--AHPDHWVIAVQWHPEER----LDDLRLFAGLVGA
MAB_0032|M.abscessus_ATCC_1997 HTEHPIHGVQFHPESILTEGGHRMLANWLTYCGAPLDEPLVRSLEAEVAA
TH_0817|M.thermoresistible__bu ANDGVVEGVRLVS-------GRAFSVQYHPEAAAG-PHDANNLFDQFIDV
: ..:. : .:: . . : : .
MSMEG_6690|M.smegmatis_MC2_155 AEQARQSRALAAAPAE--------------
Mvan_1853|M.vanbaalenii_PYR-1 AERSRENTAAVASQG---------------
Mb2884c|M.bovis_AF2122/97 ASGYAGRQSQAEPR----------------
Rv2859c|M.tuberculosis_H37Rv ASGYAGRQSQAEPR----------------
MAV_3717|M.avium_104 ARSYAGVAR---------------------
MLBr_01573|M.leprae_Br4923 ACRYANSRCVEYS-----------------
MMAR_1844|M.marinum_M ARSRRSG-----------------------
MUL_2093|M.ulcerans_Agy99 ARSRRSG-----------------------
Mflv_4064|M.gilvum_PYR-GCK AGVYATQKTQKVNR----------------
MAB_0032|M.abscessus_ATCC_1997 ALNGVTGRVAALGG----------------
TH_0817|M.thermoresistible__bu MASAASDRVCTPTSGVSRTNGDSHGDGETR