For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASYFDGVTMAGGIAVLLPPQRADADAANRVLDSLDA LVITGGKDVDPAAYGQQAHPATDEPAPIRDSWEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLP DVLGHNGHRAGNAVFSSLPVRTVPGTRLATLIGESAQVRCYHHQAVAEVGEGLVVSAWDVDGVVEALELP GENFVLAVQWHPEESLEDLRLFSAVVDAARSRRSG
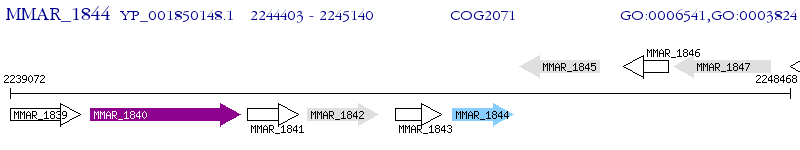
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1844 | - | - | 100% (245) | amidotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2884c | - | 1e-104 | 75.65% (230) | amidotransferase |
| M. gilvum PYR-GCK | Mflv_4064 | - | 3e-86 | 62.87% (237) | peptidase C26 |
| M. tuberculosis H37Rv | Rv2859c | - | 1e-104 | 75.65% (230) | amidotransferase |
| M. leprae Br4923 | MLBr_01573 | - | 4e-98 | 68.35% (237) | putative amidotransferase |
| M. abscessus ATCC 19977 | MAB_0032 | - | 2e-06 | 28.65% (178) | para-aminobenzoate synthase component II |
| M. avium 104 | MAV_3717 | - | 1e-101 | 73.91% (230) | class I glutamine amidotransferase, putative |
| M. smegmatis MC2 155 | MSMEG_2596 | - | 1e-97 | 72.41% (232) | peptidase C26 |
| M. thermoresistible (build 8) | TH_0484 | guaA | 4e-06 | 32.21% (149) | PROBABLE GMP SYNTHASE |
| M. ulcerans Agy99 | MUL_2093 | - | 1e-138 | 99.18% (245) | amidotransferase |
| M. vanbaalenii PYR-1 | Mvan_2279 | - | 3e-90 | 66.67% (237) | peptidase C26 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
Rv2859c|M.tuberculosis_H37Rv MDLSASRSDGGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPVSDGGDPL
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK ----------------------------------MSVS------------
Mvan_2279|M.vanbaalenii_PYR-1 ----------------------------------MNGSDPGASSRKLAQN
MSMEG_2596|M.smegmatis_MC2_155 ----------------------------------MSDSDSSE--------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0484|M.thermoresistible__bu ---VSSSADVNTSRPVLVIDFGAQYAQLIARRIREARVYSEVVPHTATVE
MMAR_1844|M.marinum_M ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASY-FDGVTM
MUL_2093|M.ulcerans_Agy99 ------MVLNAAGPGPIVGMTTYLDQAKTGVWDVSASFLPASY-FDGVTM
Mb2884c|M.bovis_AF2122/97 RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADY-FEGITM
Rv2859c|M.tuberculosis_H37Rv RPASPRLRSPLGASRPVVGLTAYLEQVRTGVWDIPAGYLPADY-FEGITM
MAV_3717|M.avium_104 -----------------MGLTAYLERVQTGIWDIPAGYLPADY-FEGVTR
MLBr_01573|M.leprae_Br4923 --------MDLSGSRPVVGLTAYLEQVHTGLWDVPAGYLPADY-FQGVAM
Mflv_4064|M.gilvum_PYR-GCK -------------SRPVIGLTTYLQQAQTGVWDVRASFLPAIY-FEGVGM
Mvan_2279|M.vanbaalenii_PYR-1 QARQRDSVSLRDRTRPVIGLTTYLQQAQTGVWDVRASFLPAIY-FEGVGM
MSMEG_2596|M.smegmatis_MC2_155 -------------LRPVIGMTTYLQQAQTGVWDVRASFLPQIY-FAGVNL
MAB_0032|M.abscessus_ATCC_1997 -----------------------MRILVVDNYDS-FVFNLVQY-LGQLGV
TH_0484|M.thermoresistible__bu EIKAKDPQAIVLSGGPASVYAEDAPRLDPGLFDLGVPVFGICYGFQAMAQ
. :* * : :
MMAR_1844|M.marinum_M AGG--------------------------IAVLLPPQRADADAANRVLDS
MUL_2093|M.ulcerans_Agy99 AGG--------------------------IAVLLPPQRADADAANRVLDS
Mb2884c|M.bovis_AF2122/97 AGG--------------------------VAVLLPPQPVDPESVGCVLDS
Rv2859c|M.tuberculosis_H37Rv AGG--------------------------VAVLLPPQPVDPESVGCVLDS
MAV_3717|M.avium_104 AGG--------------------------IAVLLPPQPVDTGIVGSLLDG
MLBr_01573|M.leprae_Br4923 AGG--------------------------IAVLLPPQPVDPEIAGLALDG
Mflv_4064|M.gilvum_PYR-GCK AGG--------------------------IASLLPPQPVDDTIAERVLDG
Mvan_2279|M.vanbaalenii_PYR-1 AGG--------------------------IAVLLPPQAADAAVADRVLDS
MSMEG_2596|M.smegmatis_MC2_155 AGG--------------------------IAVLLPPQPVDGDVADRVLDR
MAB_0032|M.abscessus_ATCC_1997 DAT--------------------------VWRNDDPRLADT---DAVAAD
TH_0484|M.thermoresistible__bu ALGGTVTRTGTREYGRTELTVLGGQLHSDLPERQPVWMSHADAVTEAPAG
: .
MMAR_1844|M.marinum_M LDALVITGGKDVDPAAYGQQAHPATDEPAPI-----------RDSWEFAL
MUL_2093|M.ulcerans_Agy99 LDALVITGGKDVDPAAYGQQAHPATDEPAPI-----------RDSWEFAL
Mb2884c|M.bovis_AF2122/97 LHALVITGGYDLDPAAYGQEPHPATDHPRPG-----------RDAWEFAL
Rv2859c|M.tuberculosis_H37Rv LHALVITGGYDLDPAAYGQEPHPATDHPRPG-----------RDAWEFAL
MAV_3717|M.avium_104 LHALVITGGYDLDPAGYGQRPHPSTDAPRTD-----------RDAWEFAL
MLBr_01573|M.leprae_Br4923 LDGLVITGGYDVDPATYGQRPHPSTDEPRTT-----------RDSWEFAL
Mflv_4064|M.gilvum_PYR-GCK IDGLIITGGRDVDPSTYGAQRHPRTDEPDTDSRA--------RDAFEFAL
Mvan_2279|M.vanbaalenii_PYR-1 LDGLIITGGRDVDPSSYGAQRHPATDEPVGDSRT--------RDAFEFAL
MSMEG_2596|M.smegmatis_MC2_155 LDGLVITGGPDVDPARYGQKPHPKTNEPALE-----------RDEWEFAL
MAB_0032|M.abscessus_ATCC_1997 FDGVLISPG-------------PGTPESAGA---------------SIAM
TH_0484|M.thermoresistible__bu FEVVATSPGSPVAAFEDPARRLAGVQYHPEVIHSPHGQQVLGRFLHEFAG
:. : : * . . .:*
MMAR_1844|M.marinum_M LRGALQRG----------LPVLG----ICRGAQVLNVAFGGTLHQHLP--
MUL_2093|M.ulcerans_Agy99 LRGALQRG----------LPVLG----ICRGAQVLNVAFGGTPHQHLP--
Mb2884c|M.bovis_AF2122/97 LRGALQRG----------MPVLG----ICRGTQVLNVALGGTLHQHLP--
Rv2859c|M.tuberculosis_H37Rv LRGALQRG----------MPVLG----ICRGTQVLNVALGGTLHQHLP--
MAV_3717|M.avium_104 LRGALQRG----------LPVLG----ICRGAQVLNVAFGGTLHQHLP--
MLBr_01573|M.leprae_Br4923 FEAALQRG----------LPVLG----ICRGAQLLNIALGGTLHQHLP--
Mflv_4064|M.gilvum_PYR-GCK VRAALRRA----------MPVLG----ICRGAQVLNVALGGTLHQHVP--
Mvan_2279|M.vanbaalenii_PYR-1 LQGALRRQ----------IPVLG----ICRGAQMLNVALGGTLHQHLP--
MSMEG_2596|M.smegmatis_MC2_155 LSGALARG----------IPVLG----VCRGAQVLNTALGGTLHQHLP--
MAB_0032|M.abscessus_ATCC_1997 VHSCAKTG----------TPLLG----VCLGHQAIGVAFGGTVDRAPE--
TH_0484|M.thermoresistible__bu IGASWTPANIAESLIEQVREQIGDGRAICGLSGGVDSAVAAALVQRAIGD
. .. :* :* :. *...: :
MMAR_1844|M.marinum_M ----DVLGHNGHRAGNAVFSSLPVRTVPGTRLATLIGES-----------
MUL_2093|M.ulcerans_Agy99 ----DVLGHNGHRAGNAVFSSLPVRTVPGTRLATLIGES-----------
Mb2884c|M.bovis_AF2122/97 ----DILGHSGHRAGNGVFTRLPVHTASGTRLAELIGES-----------
Rv2859c|M.tuberculosis_H37Rv ----DILGHSGHRAGNGVFTRLPVHTASGTRLAELIGES-----------
MAV_3717|M.avium_104 ----DVLGHGGHRAGNGVFSTLPVRTVAGTRLAALLGET-----------
MLBr_01573|M.leprae_Br4923 ----EVIGHSKHWVGNAVFNNLLVRTVPGTRLAAVLGEF-----------
Mflv_4064|M.gilvum_PYR-GCK ----DVVGHTRHQQGNAVFTTSSIATVPGSRVAALVGPD-----------
Mvan_2279|M.vanbaalenii_PYR-1 ----DVVGHTRHQQGNAVFTTSSITTVPGTRVAALVGPD-----------
MSMEG_2596|M.smegmatis_MC2_155 ----DVIGHTHHQKGDAVFGTSDVRTVSGTRLASLIGES-----------
MAB_0032|M.abscessus_ATCC_1997 ----LLHGKTSTVEHEDAGVLRGLPNPFTATRYHSLTIL-----------
TH_0484|M.thermoresistible__bu RLTCVFVDHGLLRAGERAQVQRDFVAATGAKLVTVDTSERFLEALSGVTN
. .: : . . :
MMAR_1844|M.marinum_M ----------AQVRCYHHQAVAEVGEGLVVSAWDVDGVVEALELP-----
MUL_2093|M.ulcerans_Agy99 ----------AQVRCYHHQAVAEVGEGLVVSAFDVDGVVEALELP-----
Mb2884c|M.bovis_AF2122/97 ----------ADVPCYHHQAIDQVGEGLVVSAVDVDGVIEALELP-----
Rv2859c|M.tuberculosis_H37Rv ----------ADVPCYHHQAIDQVGEGLVVSAVDVDGVIEALELP-----
MAV_3717|M.avium_104 ----------VDAPCYHHQAIDKLGDGLVVSAWDPDGVVEAVELP-----
MLBr_01573|M.leprae_Br4923 ----------VEARCYHHQSIDKLGDGLVVSAWDADGVVEAVELP-----
Mflv_4064|M.gilvum_PYR-GCK ----------TEAQCYHHQAIDRLGDGLVVSASDTDGVIEAVEVDLS---
Mvan_2279|M.vanbaalenii_PYR-1 ----------IEAQCYHHQAVDRLGDGLIVSARGVDGVVEAVELDPA---
MSMEG_2596|M.smegmatis_MC2_155 ----------SDAQCYHHQAIDRLGEGLVVSARDTDGVIEAVEVP-----
MAB_0032|M.abscessus_ATCC_1997 ----------PDTVPEGLRVIARTASGVIMGVMHTEHPIHGVQFHPESIL
TH_0484|M.thermoresistible__bu PEGKRKIIGREFIRAFEGAVREALGENAAANDDEVEFLVQGTLYPDVVE-
... . : :..
MMAR_1844|M.marinum_M ---GENFVLAVQWHPEE---------SLED--LRLFSAVVDAARSRRSG-
MUL_2093|M.ulcerans_Agy99 ---GENFVLAVQWHPEE---------SLED--LRLFSAVVDAARSRRSG-
Mb2884c|M.bovis_AF2122/97 ---GDTFVLAVQWHPEK---------SLDD--LRLFKALVDAASGYAGRQ
Rv2859c|M.tuberculosis_H37Rv ---GDTFVLAVQWHPEK---------SLDD--LRLFKALVDAASGYAGRQ
MAV_3717|M.avium_104 ---GDAFVLAVQWHPEQ---------SLHD--LRLFTAIVDAARSYAGVA
MLBr_01573|M.leprae_Br4923 ---GDAFVLGVQWHPEK---------ALSD--LRLFTAIVDAACRYANSR
Mflv_4064|M.gilvum_PYR-GCK -AHPDHWVIAVQWHPEE---------RLDD--LRLFAGLVGAAGVYATQK
Mvan_2279|M.vanbaalenii_PYR-1 -TRSDGWAVAVQWHPEE---------RLDD--LRLFAGLVSAAGDYAWHK
MSMEG_2596|M.smegmatis_MC2_155 ---GEAFVLGVQWHPEE---------RLDD--LRLFRAVVEAARTYATER
MAB_0032|M.abscessus_ATCC_1997 TEGGHRMLANWLTYCGA---------PLDEPLVRSLEAEVAAALNGVTGR
TH_0484|M.thermoresistible__bu -SGGGAGTANIKSHHNVGGLPDDLKFKLVEPLRLLFKDEVRAVGRELGLP
: * : : * *.
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 SQAEPR--------------------------------------------
Rv2859c|M.tuberculosis_H37Rv SQAEPR--------------------------------------------
MAV_3717|M.avium_104 R-------------------------------------------------
MLBr_01573|M.leprae_Br4923 CVEYS---------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK TQKVNR--------------------------------------------
Mvan_2279|M.vanbaalenii_PYR-1 IEKVS---------------------------------------------
MSMEG_2596|M.smegmatis_MC2_155 VS------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 VAALGG--------------------------------------------
TH_0484|M.thermoresistible__bu EEIVARQPFPGPGLAIRIVGEVTAERLETLRRADAIVREELTAAGLDHQI
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mb2884c|M.bovis_AF2122/97 --------------------------------------------------
Rv2859c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2279|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2596|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0484|M.thermoresistible__bu WQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYDVL
MMAR_1844|M.marinum_M --------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------
Mb2884c|M.bovis_AF2122/97 --------------------------------
Rv2859c|M.tuberculosis_H37Rv --------------------------------
MAV_3717|M.avium_104 --------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------
Mvan_2279|M.vanbaalenii_PYR-1 --------------------------------
MSMEG_2596|M.smegmatis_MC2_155 --------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------
TH_0484|M.thermoresistible__bu ERISTRITNEVAEVNRVVLDITSKPPATIEWE