For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DLSGSRPVVGLTAYLEQVHTGLWDVPAGYLPADYFQGVAMAGGIAVLLPPQPVDPEIAGLALDGLDGLVI TGGYDVDPATYGQRPHPSTDEPRTTRDSWEFALFEAALQRGLPVLGICRGAQLLNIALGGTLHQHLPEVI GHSKHWVGNAVFNNLLVRTVPGTRLAAVLGEFVEARCYHHQSIDKLGDGLVVSAWDADGVVEAVELPGDA FVLGVQWHPEKALSDLRLFTAIVDAACRYANSRCVEYS*
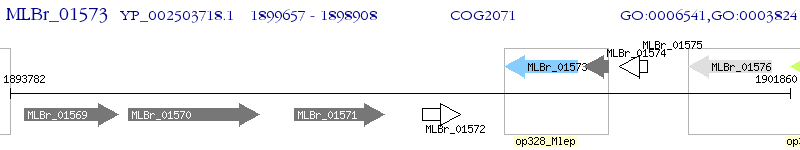
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01573 | - | - | 100% (249) | putative amidotransferase |
| M. leprae Br4923 | MLBr_00535 | carA | 5e-05 | 31.15% (122) | carbamoyl phosphate synthase small subunit |
| M. leprae Br4923 | MLBr_00015 | pabA | 5e-05 | 30.41% (148) | para-aminobenzoate synthase component II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2884c | - | 1e-104 | 71.55% (239) | amidotransferase |
| M. gilvum PYR-GCK | Mflv_4064 | - | 3e-89 | 63.82% (246) | peptidase C26 |
| M. tuberculosis H37Rv | Rv2859c | - | 1e-104 | 71.55% (239) | amidotransferase |
| M. abscessus ATCC 19977 | MAB_0032 | - | 2e-06 | 30.07% (143) | para-aminobenzoate synthase component II |
| M. marinum M | MMAR_1844 | - | 1e-97 | 68.35% (237) | amidotransferase |
| M. avium 104 | MAV_3717 | - | 1e-108 | 79.31% (232) | class I glutamine amidotransferase, putative |
| M. smegmatis MC2 155 | MSMEG_2596 | - | 2e-96 | 67.90% (243) | peptidase C26 |
| M. thermoresistible (build 8) | TH_0817 | carA | 2e-05 | 28.08% (146) | PROBABLE CARBAMOYL-PHOSPHATE SYNTHASE SMALL CHAIN CARA |
| M. ulcerans Agy99 | MUL_2093 | - | 8e-96 | 67.51% (237) | amidotransferase |
| M. vanbaalenii PYR-1 | Mvan_2279 | - | 1e-87 | 62.55% (243) | peptidase C26 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2884c|M.bovis_AF2122/97 --------------------------------------------------
Rv2859c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2279|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2596|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0817|M.thermoresistible__bu MTMTASTQATTAATRAVLVLEDGRVFTGVAFGAVGETLGEAVFTTGMSGY
Mb2884c|M.bovis_AF2122/97 -----------------------MDLSASRSDGGDPLRPASPRLRSPVSD
Rv2859c|M.tuberculosis_H37Rv -----------------------MDLSASRSDGGDPLRPASPRLRSPVSD
MAV_3717|M.avium_104 --------------------------------------------------
MLBr_01573|M.leprae_Br4923 --------------------------------------------------
MMAR_1844|M.marinum_M --------------------------------------------------
MUL_2093|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4064|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2279|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2596|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0032|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0817|M.thermoresistible__bu QEALTDPSFHRQIVIATAPHIGNTGWNDEDSESRGDKIWVAGYAVRDPSP
Mb2884c|M.bovis_AF2122/97 GGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPLGASRPVVGLTAYLEQV
Rv2859c|M.tuberculosis_H37Rv GGDPLRPASPRLRSPVSDGGDPLRPASPRLRSPLGASRPVVGLTAYLEQV
MAV_3717|M.avium_104 ----------------------------------------MGLTAYLERV
MLBr_01573|M.leprae_Br4923 -------------------------------MDLSGSRPVVGLTAYLEQV
MMAR_1844|M.marinum_M -----------------------------MVLNAAGPGPIVGMTTYLDQA
MUL_2093|M.ulcerans_Agy99 -----------------------------MVLNAAGPGPIVGMTTYLDQA
Mflv_4064|M.gilvum_PYR-GCK -------MSVS-------------------------SRPVIGLTTYLQQA
Mvan_2279|M.vanbaalenii_PYR-1 -------MNGSDPGASSRKLAQNQARQRDSVSLRDRTRPVIGLTTYLQQA
MSMEG_2596|M.smegmatis_MC2_155 -------MSDSDSSE---------------------LRPVIGMTTYLQQA
MAB_0032|M.abscessus_ATCC_1997 ----------------------------------------------MRIL
TH_0817|M.thermoresistible__bu RASNWRATRTLEEELVRQNIVGIAGIDTRAVVRHLRTRGTMKAGVFSGPA
Mb2884c|M.bovis_AF2122/97 RTGVWDIPAGYLPADYFEGITMAGGVAV----LLPPQ-------------
Rv2859c|M.tuberculosis_H37Rv RTGVWDIPAGYLPADYFEGITMAGGVAV----LLPPQ-------------
MAV_3717|M.avium_104 QTGIWDIPAGYLPADYFEGVTRAGGIAV----LLPPQ-------------
MLBr_01573|M.leprae_Br4923 HTGLWDVPAGYLPADYFQGVAMAGGIAV----LLPPQ-------------
MMAR_1844|M.marinum_M KTGVWDVSASFLPASYFDGVTMAGGIAV----LLPPQ-------------
MUL_2093|M.ulcerans_Agy99 KTGVWDVSASFLPASYFDGVTMAGGIAV----LLPPQ-------------
Mflv_4064|M.gilvum_PYR-GCK QTGVWDVRASFLPAIYFEGVGMAGGIAS----LLPPQ-------------
Mvan_2279|M.vanbaalenii_PYR-1 QTGVWDVRASFLPAIYFEGVGMAGGIAV----LLPPQ-------------
MSMEG_2596|M.smegmatis_MC2_155 QTGVWDVRASFLPQIYFAGVNLAGGIAV----LLPPQ-------------
MAB_0032|M.abscessus_ATCC_1997 VVDNYDS-FVFNLVQYLGQLGVDATVWR----NDDPR-------------
TH_0817|M.thermoresistible__bu LADIDELVTRVREQPSMLGANLSGEVTTPEPYVLEPEGPHRFVIAAIDLG
.. : : . : *.
Mb2884c|M.bovis_AF2122/97 PVDPESVGCVLDSLHALVITGGYDLDPAAYGQEPHPATDHPRPG---RDA
Rv2859c|M.tuberculosis_H37Rv PVDPESVGCVLDSLHALVITGGYDLDPAAYGQEPHPATDHPRPG---RDA
MAV_3717|M.avium_104 PVDTGIVGSLLDGLHALVITGGYDLDPAGYGQRPHPSTDAPRTD---RDA
MLBr_01573|M.leprae_Br4923 PVDPEIAGLALDGLDGLVITGGYDVDPATYGQRPHPSTDEPRTT---RDS
MMAR_1844|M.marinum_M RADADAANRVLDSLDALVITGGKDVDPAAYGQQAHPATDEPAPI---RDS
MUL_2093|M.ulcerans_Agy99 RADADAANRVLDSLDALVITGGKDVDPAAYGQQAHPATDEPAPI---RDS
Mflv_4064|M.gilvum_PYR-GCK PVDDTIAERVLDGIDGLIITGGRDVDPSTYGAQRHPRTDEPDTDSRARDA
Mvan_2279|M.vanbaalenii_PYR-1 AADAAVADRVLDSLDGLIITGGRDVDPSSYGAQRHPATDEPVGDSRTRDA
MSMEG_2596|M.smegmatis_MC2_155 PVDGDVADRVLDRLDGLVITGGPDVDPARYGQKPHPKTNEPALE---RDE
MAB_0032|M.abscessus_ATCC_1997 LADT---DAVAADFDGVLISPG-------------PGTPESAGA------
TH_0817|M.thermoresistible__bu IKTNTPRNFVQRGVRTHVMPAGTTFDELRDLKPDGVFLSNGPGDP-ATAD
. ::. *
Mb2884c|M.bovis_AF2122/97 WEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPDILGHSGHRAG
Rv2859c|M.tuberculosis_H37Rv WEFALLRGALQRGMPVLGICRGTQVLNVALGGTLHQHLPDILGHSGHRAG
MAV_3717|M.avium_104 WEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPDVLGHGGHRAG
MLBr_01573|M.leprae_Br4923 WEFALFEAALQRGLPVLGICRGAQLLNIALGGTLHQHLPEVIGHSKHWVG
MMAR_1844|M.marinum_M WEFALLRGALQRGLPVLGICRGAQVLNVAFGGTLHQHLPDVLGHNGHRAG
MUL_2093|M.ulcerans_Agy99 WEFALLRGALQRGLPVLGICRGAQVLNVAFGGTPHQHLPDVLGHNGHRAG
Mflv_4064|M.gilvum_PYR-GCK FEFALVRAALRRAMPVLGICRGAQVLNVALGGTLHQHVPDVVGHTRHQQG
Mvan_2279|M.vanbaalenii_PYR-1 FEFALLQGALRRQIPVLGICRGAQMLNVALGGTLHQHLPDVVGHTRHQQG
MSMEG_2596|M.smegmatis_MC2_155 WEFALLSGALARGIPVLGVCRGAQVLNTALGGTLHQHLPDVIGHTHHQKG
MAB_0032|M.abscessus_ATCC_1997 -SIAMVHSCAKTGTPLLGVCLGHQAIGVAFGGTVDRAPELLHGKTSTVEH
TH_0817|M.thermoresistible__bu HVVAVTRSVLEAGIPLFGICFGNQILGRALGRSTYKMVFGHRGHNVPVID
.*: . *::*:* * * :. *:* : : *:
Mb2884c|M.bovis_AF2122/97 NGVFTRLPVHTASGTRLAELIGESADVPCYHHQAIDQVG-EGLVVSAVDV
Rv2859c|M.tuberculosis_H37Rv NGVFTRLPVHTASGTRLAELIGESADVPCYHHQAIDQVG-EGLVVSAVDV
MAV_3717|M.avium_104 NGVFSTLPVRTVAGTRLAALLGETVDAPCYHHQAIDKLG-DGLVVSAWDP
MLBr_01573|M.leprae_Br4923 NAVFNNLLVRTVPGTRLAAVLGEFVEARCYHHQSIDKLG-DGLVVSAWDA
MMAR_1844|M.marinum_M NAVFSSLPVRTVPGTRLATLIGESAQVRCYHHQAVAEVG-EGLVVSAWDV
MUL_2093|M.ulcerans_Agy99 NAVFSSLPVRTVPGTRLATLIGESAQVRCYHHQAVAEVG-EGLVVSAFDV
Mflv_4064|M.gilvum_PYR-GCK NAVFTTSSIATVPGSRVAALVGPDTEAQCYHHQAIDRLG-DGLVVSASDT
Mvan_2279|M.vanbaalenii_PYR-1 NAVFTTSSITTVPGTRVAALVGPDIEAQCYHHQAVDRLG-DGLIVSARGV
MSMEG_2596|M.smegmatis_MC2_155 DAVFGTSDVRTVSGTRLASLIGESSDAQCYHHQAIDRLG-EGLVVSARDT
MAB_0032|M.abscessus_ATCC_1997 EDAGVLRGLPNPFTATRYHSLTILPDTVPEGLRVIARTA-SGVIMGVMHT
TH_0817|M.thermoresistible__bu HRTGRVAITAQNHGFALEGEAGEVFDTPFGRAEISHTCANDGVVEGVRLV
. . :. . . .*:: ..
Mb2884c|M.bovis_AF2122/97 DGVIEALELP--------GDTFVLAVQWHPEKSLDD--LRLFKALVDAAS
Rv2859c|M.tuberculosis_H37Rv DGVIEALELP--------GDTFVLAVQWHPEKSLDD--LRLFKALVDAAS
MAV_3717|M.avium_104 DGVVEAVELP--------GDAFVLAVQWHPEQSLHD--LRLFTAIVDAAR
MLBr_01573|M.leprae_Br4923 DGVVEAVELP--------GDAFVLGVQWHPEKALSD--LRLFTAIVDAAC
MMAR_1844|M.marinum_M DGVVEALELP--------GENFVLAVQWHPEESLED--LRLFSAVVDAAR
MUL_2093|M.ulcerans_Agy99 DGVVEALELP--------GENFVLAVQWHPEESLED--LRLFSAVVDAAR
Mflv_4064|M.gilvum_PYR-GCK DGVIEAVEVDLS----AHPDHWVIAVQWHPEERLDD--LRLFAGLVGAAG
Mvan_2279|M.vanbaalenii_PYR-1 DGVVEAVELDPA----TRSDGWAVAVQWHPEERLDD--LRLFAGLVSAAG
MSMEG_2596|M.smegmatis_MC2_155 DGVIEAVEVP--------GEAFVLGVQWHPEERLDD--LRLFRAVVEAAR
MAB_0032|M.abscessus_ATCC_1997 EHPIHGVQFHPESILTEGGHRMLANWLTYCGAPLDEPLVRSLEAEVAAAL
TH_0817|M.thermoresistible__bu SGRAFSVQYHPEAAAGPHDANNLFDQFIDVMASAAS--DRVCTPTSGVSR
. .:: . * .:
Mb2884c|M.bovis_AF2122/97 GYAGRQSQAEPR
Rv2859c|M.tuberculosis_H37Rv GYAGRQSQAEPR
MAV_3717|M.avium_104 SYAGVAR-----
MLBr_01573|M.leprae_Br4923 RYANSRCVEYS-
MMAR_1844|M.marinum_M SRRSG-------
MUL_2093|M.ulcerans_Agy99 SRRSG-------
Mflv_4064|M.gilvum_PYR-GCK VYATQKTQKVNR
Mvan_2279|M.vanbaalenii_PYR-1 DYAWHKIEKVS-
MSMEG_2596|M.smegmatis_MC2_155 TYATERVS----
MAB_0032|M.abscessus_ATCC_1997 NGVTGRVAALGG
TH_0817|M.thermoresistible__bu TNGDSHGDGETR