For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRQIRSLVVAAAAAAAILATVAPGGSAHAQPAEVKNVVLVHGAFADGSGWRGVYDELTARGYRVTIVQNP LTSLADDVAATKRVLDQQDGPTLLVGHSYGGTVITEAGVDPKVTGLVYVSGLAPDAGETTSQQYADYPTP PSFVIETGPDGFGFINREKFKAGFAADTSDEVAAFLRDSQVPINMAIFGDTLTQAAWRTKPSWAVVATQD EAIDPRMLRQMAQRIGAQITDVEGSHVVFLTNPRAVADVIDRAAKESGRA*
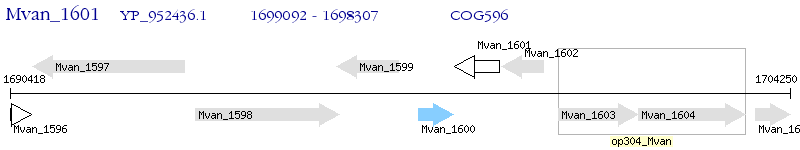
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1601 | - | - | 100% (261) | hypothetical protein Mvan_1601 |
| M. vanbaalenii PYR-1 | Mvan_4902 | - | 7e-51 | 44.40% (259) | hypothetical protein Mvan_4902 |
| M. vanbaalenii PYR-1 | Mvan_4177 | - | 2e-09 | 27.20% (250) | esterase EstC, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2476 | - | 2e-08 | 26.48% (253) | esterase EstC, putative |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0768 | - | 1e-47 | 45.45% (220) | hypothetical protein MAB_0768 |
| M. marinum M | MMAR_2050 | - | 5e-07 | 41.38% (87) | hypothetical protein MMAR_2050 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_5107 | - | 8e-48 | 46.33% (218) | secreted protein |
| M. thermoresistible (build 8) | TH_2893 | - | 8e-25 | 31.00% (229) | PUTATIVE putative conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3299 | - | 3e-07 | 41.38% (87) | hypothetical protein MUL_3299 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1601|M.vanbaalenii_PYR-1 ---------------------------------MNRQIRSLVVAAAAAAA
MSMEG_5107|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0768|M.abscessus_ATCC_1997 --------------------------------------------------
TH_2893|M.thermoresistible__bu --------------------------------------------------
MMAR_2050|M.marinum_M MRPTLAAAALAGASGLVGWGGARALARRIERNPDPFPRERLLAEPQGERV
MUL_3299|M.ulcerans_Agy99 MRPTLAAAALAGASGLVGWGGARALARRIERNPDPFPRERLLAEPQGERV
Mflv_2476|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1601|M.vanbaalenii_PYR-1 ILATVAPGGSAHAQPAEVKNVVLVHGAFADGSGWRGVYDELTARGYR-VT
MSMEG_5107|M.smegmatis_MC2_155 ------------------MNIVLVHGAFVDGSTWRVVYDLLVDEGHR-VV
MAB_0768|M.abscessus_ATCC_1997 --------------MSSKPTIVLVHGFWGGAAHWGNVIVELHGRGFEDLH
TH_2893|M.thermoresistible__bu ----------------MAKPILLVHGAFSGSWVWDQVVAELASCGVQART
MMAR_2050|M.marinum_M AITRPDGTVLHALVAGRGTPVILVHGYCASLTEWNFVWDELLARGLRVIA
MUL_3299|M.ulcerans_Agy99 AITRPDGTVLHALVAGRGTPVILVHGYCASLTEWNFVWDELLARGLRVIA
Mflv_2476|M.gilvum_PYR-GCK ------------------MRFILVHGGFHAAWCWERTAAALRALGHDVTA
.:**** * . * *
Mvan_1601|M.vanbaalenii_PYR-1 IVQN------------PLTSLADDVAATKRVLDQQDGPTLLVGHSYGGTV
MSMEG_5107|M.smegmatis_MC2_155 VVQT------------PNLSLSGDVEATRRVLDSLVGSVVLVGHSCGGAV
MAB_0768|M.abscessus_ATCC_1997 AVEN------------PLTSLADDAARTRQMVQQIDGPVLLVGHSYGGAV
TH_2893|M.thermoresistible__bu VELSSR---------KPDGTLARDAHVVRDALKQFDQPAVLVGHSYGGAV
MMAR_2050|M.marinum_M FDQRGHGRSTLGAEGIGSEPMADDLAAVLQHFDIRDG--VLVGHSMGGFV
MUL_3299|M.ulcerans_Agy99 FDQRGHGRSTLGAEGIGSEPMADDLAAVLQHFDLRDG--VLVGHSMGGFV
Mflv_2476|M.gilvum_PYR-GCK VDLPGHG-----ALADQESTLANRRDAIVAAMQAGDEKCVLVGHSGGGFD
.:: .. :***** **
Mvan_1601|M.vanbaalenii_PYR-1 ITEAG-VDPKVTG----LVYVSGLAPDAGETTSQQYADYPTPPS---FVI
MSMEG_5107|M.smegmatis_MC2_155 ITEAG-NHDSVSA----LVYIAAFVPDEGES-VESLGGHPGVQG---SPI
MAB_0768|M.abscessus_ATCC_1997 ITEAG-DLPNVAG----LVYVAAFAPDAGESPGQLTEQLPPAAA---ANL
TH_2893|M.thermoresistible__bu ITEASADNDHVAH----LIYVCAALPQAGESVSDVLARDPDPQGDLAPAL
MMAR_2050|M.marinum_M SIRALLDHGALAQRLRAVVLFATWAGRVLDGAPQNRLQIPLLERGILQRL
MUL_3299|M.ulcerans_Agy99 SIRALLDHGALAQRLRAVVLFATWAGRVLDGAPQNRLQIPLLERGILQRL
Mflv_2476|M.gilvum_PYR-GCK ATLAADARPDLVS---HITYLAAALPREGRTYPEAMAMRDDDNGPVELGE
* : : .. :
Mvan_1601|M.vanbaalenii_PYR-1 ETGPDGFGFINREKFKAG--------------FAADTSDEVAAFLRDSQV
MSMEG_5107|M.smegmatis_MC2_155 VPFSGGFLLQDRAKFHSS--------------FGADLPTADAAFLADSQI
MAB_0768|M.abscessus_ATCC_1997 VPDSDGYLWIKQDKFRES--------------FAQDLPEDAALVMAVTQK
TH_2893|M.thermoresistible__bu EVREDGTATMKRDAARQA--------------LFNDATEEQFESVVDKLG
MMAR_2050|M.marinum_M LRSRTIAVLFGAAQCGAR--------------PSPAMISVFLEWFHQHLD
MUL_3299|M.ulcerans_Agy99 LRSRTIAVLFGAAQCGAR--------------PSPAMISG-IPRVVPSTS
Mflv_2476|M.gilvum_PYR-GCK DFDGDVGEMLGYLHFDDTGAMKFADFDGAWKYFYHDCDEETARWAFERLG
Mvan_1601|M.vanbaalenii_PYR-1 PINM-AIFGDTLTQAAWRTKP-----------SWAVVATQDEAIDPRMLR
MSMEG_5107|M.smegmatis_MC2_155 PWST-EAMTSPVTDPAWRRRP-----------CWYLIATEDRMIPLAAQR
MAB_0768|M.abscessus_ATCC_1997 APLA-STFGDAITAPAWRNKP-----------SWYQVSTQDRMINPENER
TH_2893|M.thermoresistible__bu PHAM-GTLSEPVTGLGWQQHP-----------ATYVIALRDQMFSVALQE
MMAR_2050|M.marinum_M RHGP-HLPIVRA--FSREDRYPRLAEISVP--AVVMVGSSDRTTPPSHSH
MUL_3299|M.ulcerans_Agy99 RPAR-TAPTHRAGVLARGSLSTAGRDLGAGGGDGGLLGSHHPAQPLAPAG
Mflv_2476|M.gilvum_PYR-GCK PERFGDTTVTPVSVPNFWAADLP---------RSFIVCEQDQAMPRWLAD
: .
Mvan_1601|M.vanbaalenii_PYR-1 QMAQRIG-----AQITDVEGSHVVFLTNPRAVADVIDRAAKESGRA----
MSMEG_5107|M.smegmatis_MC2_155 VMAQRAG-----AATVEVAASHAVYMSQPRAVAALIGQAVHV--------
MAB_0768|M.abscessus_ATCC_1997 RMAQRIKP----RKTIELDASHASLASQPVAIADLIAEAAGDIA------
TH_2893|M.thermoresistible__bu EFASHVG------TVAKIDTGHGPMATRPAELAEIIATAAR---------
MMAR_2050|M.marinum_M RLAAGIPG---ARLVT--VPGAGHLLN--WEAVDELAEVVESFAAQR---
MUL_3299|M.ulcerans_Agy99 RRNPGCPAGHRARRGTPAELGGGRRAGRSRRILCGAAVLMRGHAAGVWR-
Mflv_2476|M.gilvum_PYR-GCK TVARRLG-----VDQLTIDASHSPFLSRPREFADLLVHATTTTPVAPLRP
.
Mvan_1601|M.vanbaalenii_PYR-1 -
MSMEG_5107|M.smegmatis_MC2_155 -
MAB_0768|M.abscessus_ATCC_1997 -
TH_2893|M.thermoresistible__bu -
MMAR_2050|M.marinum_M -
MUL_3299|M.ulcerans_Agy99 -
Mflv_2476|M.gilvum_PYR-GCK L