For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VESATPRPVLVVDFGAQYAQLIARRVREARVFSEVIPHTTSVEEIKARNPRAVVLSGGPASVYADGAPQL DPALFDLDVPVFGICYGFQAMAQALGGTVAHTGTSEYGRTELKVAGGQLHTDLPETQPVWMSHGDAVTAA PEGFEVVATSAGAPVAAFENRARRLAGVQYHPEVMHSPHGQQVLSRFLHEFAGIDATWTPANIAEALVEQ VRTQIGDGRAICGLSGGVDSAVAAALVQRAIGDRLTCVFVDHGLLRSGERAQVQRDFVAATGANLVTVDV ADRFLEALSGVTNPEGKRKIIGREFIRAFEGAVRDALESQDGSDVPVEFLVQGTLYPDVVESGGGSGTAN IKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRAVGRELGLPEEIVARQPFPGPGLGIRIVGEVTAERLDTL RRADAIAREELTSAGLDHQIWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEVL ERISTRITNEVREVNRVVLDVTSKPPGTIEWE
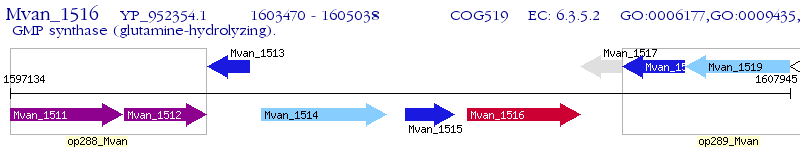
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1516 | guaA | - | 100% (522) | bifunctional GMP synthase/glutamine amidotransferase protein |
| M. vanbaalenii PYR-1 | Mvan_1852 | - | 8e-08 | 33.93% (112) | peptidase C26 |
| M. vanbaalenii PYR-1 | Mvan_2980 | - | 2e-07 | 28.06% (139) | glutamine amidotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3429c | guaA | 0.0 | 88.12% (522) | GMP synthase |
| M. gilvum PYR-GCK | Mflv_4903 | guaA | 0.0 | 94.44% (522) | bifunctional GMP synthase/glutamine amidotransferase protein |
| M. tuberculosis H37Rv | Rv3396c | guaA | 0.0 | 88.12% (522) | GMP synthase |
| M. leprae Br4923 | MLBr_00395 | guaA | 0.0 | 86.68% (518) | GMP synthase |
| M. abscessus ATCC 19977 | MAB_3718c | - | 0.0 | 86.07% (517) | GMP synthase |
| M. marinum M | MMAR_1148 | guaA | 0.0 | 89.92% (516) | GMP synthase, GuaA |
| M. avium 104 | MAV_4348 | guaA | 0.0 | 88.91% (514) | bifunctional GMP synthase/glutamine amidotransferase protein |
| M. smegmatis MC2 155 | MSMEG_1610 | guaA | 0.0 | 89.66% (522) | bifunctional GMP synthase/glutamine amidotransferase protein |
| M. thermoresistible (build 8) | TH_0484 | guaA | 0.0 | 88.25% (519) | PROBABLE GMP SYNTHASE |
| M. ulcerans Agy99 | MUL_0913 | guaA | 0.0 | 89.92% (516) | GMP synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1516|M.vanbaalenii_PYR-1 --MES--------ATPRPVLVVDFGAQYAQLIARRVREARVFSEVIPHTT
Mflv_4903|M.gilvum_PYR-GCK --MES--------AHPRPVLIVDFGAQYAQLIARRVREARVFSEVIPHTA
MSMEG_1610|M.smegmatis_MC2_155 --MAS--------ESPRPVLVIDYGAQYAQLIARRVREARVYSEVVPHTA
TH_0484|M.thermoresistible__bu --VSSSADV----NTSRPVLVIDFGAQYAQLIARRIREARVYSEVVPHTA
MLBr_00395|M.leprae_Br4923 --MADSAGLDQPEPSPRPVLVVDFGAQYAQLIARRVREARVFSEVIPHTT
MAV_4348|M.avium_104 ------------------MLVVDFGAQYAQLIARRVREARVFSEVIPHTA
Mb3429c|M.bovis_AF2122/97 --MVQPADIDVPETPARPVLVVDFGAQYAQLIARRVREARVFSEVIPHTA
Rv3396c|M.tuberculosis_H37Rv --MVQPADIDVPETPARPVLVVDFGAQYAQLIARRVREARVFSEVIPHTA
MMAR_1148|M.marinum_M --MAEPVDLDVAGPVGRPVLVVDFGAQYAQLIARRVREARVFSEVIPHTT
MUL_0913|M.ulcerans_Agy99 MPVAEPVDLDVAGPVGRPVLVVDFGAQYAQLIARRVREARVFSEVIPHTT
MAB_3718c|M.abscessus_ATCC_199 --MAQTEQ-----HGDRPVLVIDFGAQYAQLIARRVREARVFSEVIPHSA
:*::*:***********:*****:***:**::
Mvan_1516|M.vanbaalenii_PYR-1 SVEEIKARNPRAVVLSGGPASVYADGAPQLDPALFDLDVPVFGICYGFQA
Mflv_4903|M.gilvum_PYR-GCK SIEEIKARDPRAVVLSGGPSSVYADGAPQLDPALFDLDIPVFGICYGFQA
MSMEG_1610|M.smegmatis_MC2_155 GVEEIKAKDPQAIVLSGGPASVYSEGAPQLDPGLFDLDVPVFGICYGFQA
TH_0484|M.thermoresistible__bu TVEEIKAKDPQAIVLSGGPASVYAEDAPRLDPGLFDLGVPVFGICYGFQA
MLBr_00395|M.leprae_Br4923 SIEEITARDPLAIVLSGGPASVYTEGAPQLDPALFDLGLPVFGICYGFQA
MAV_4348|M.avium_104 TIEEIKARQPRALVLSGGPSSVYEPGAPQLDPAVFDLGVPVFGICYGFQA
Mb3429c|M.bovis_AF2122/97 SIEEIRARQPVALVLSGGPASVYADGAPKLDPALLDLGVPVLGICYGFQA
Rv3396c|M.tuberculosis_H37Rv SIEEIRARQPVALVLSGGPASVYADGAPKLDPALLDLGVPVLGICYGFQA
MMAR_1148|M.marinum_M SIEEIKARDPLALVLSGGPASVYAPGAPQLDPALFDLGLPVFGICYGFQV
MUL_0913|M.ulcerans_Agy99 SIEEIKARDPLALVLSGGPASVYAPGAPQLDPALFDLGLPVFGICYGFQV
MAB_3718c|M.abscessus_ATCC_199 SVEEIKGRNPRAIVLSGGPSSVYEEGAPQLDPGVFDLDVPVFGICYGFQA
:*** .::* *:******:*** .**:***.::**.:**:*******.
Mvan_1516|M.vanbaalenii_PYR-1 MAQALGGTVAHTGTSEYGRTELKVAGGQLHTDLPETQPVWMSHGDAVTAA
Mflv_4903|M.gilvum_PYR-GCK MAQALGGTVARTGTSEYGRTELKVAGGELHSDLPEVQPVWMSHGDAVTEA
MSMEG_1610|M.smegmatis_MC2_155 MAQALGGTVERTGTSEFGRTELQVTGGDLHSELPATQPVWMSHGDAVTAA
TH_0484|M.thermoresistible__bu MAQALGGTVTRTGTREYGRTELTVLGGQLHSDLPERQPVWMSHADAVTEA
MLBr_00395|M.leprae_Br4923 MAQVLGGTVARTKTSEYGRTELKVLGGDLHSGLPGVQPVWMSHGDAVTAA
MAV_4348|M.avium_104 MAQALGGTVAHTGTSEYGRTELKVLGGELHSGLPGVQPVWMSHGDAVTAA
Mb3429c|M.bovis_AF2122/97 MAQALGGIVAHTGTREYGRTELKVLGGKLHSDLPEVQPVWMSHGDAVTAA
Rv3396c|M.tuberculosis_H37Rv MAQALGGIVAHTGTREYGRTELKVLGGKLHSDLPEVQPVWMSHGDAVTAA
MMAR_1148|M.marinum_M MAQALGGTVAHTGTSEYGRTELKVLGGDLHSDLPDIQPVWMSHGDAVTAA
MUL_0913|M.ulcerans_Agy99 MAQALGGTVAHTGTSEYGRTELKVLGGDLHSDLPDIQPVWMSHGDAVTAA
MAB_3718c|M.abscessus_ATCC_199 MAQVLGGTVAHTGTSEYGRTELTVAGGDLHEGLPDVQPVWMSHGDAVTEA
***.*** * :* * *:***** * **.** ** *******.**** *
Mvan_1516|M.vanbaalenii_PYR-1 PEGFEVVATSAGAPVAAFENRARRLAGVQYHPEVMHSPHGQQVLSRFLHE
Mflv_4903|M.gilvum_PYR-GCK PSGFEVVATSVGAPVAAFENRARRLAGVQYHPEVMHSPHGQQVLSRFLHE
MSMEG_1610|M.smegmatis_MC2_155 PEGFDVVASSAGAPVAGFENRARRLAGVQYHPEVLHSPHGQQVLSRFLHD
TH_0484|M.thermoresistible__bu PAGFEVVATSPGSPVAAFEDPARRLAGVQYHPEVIHSPHGQQVLGRFLHE
MLBr_00395|M.leprae_Br4923 PDGFDVAASSSGAPVAAFENRDRRLAGVQYHPEVMHTPHGQQVLSRFLHD
MAV_4348|M.avium_104 PDGFEVVASSPGAVVAAFENRARRLAGVQYHPEVMHTPHGQQVLSRFLHD
Mb3429c|M.bovis_AF2122/97 PDGFDVVASSAGAPVAAFEAFDRRLAGVQYHPEVMHTPHGQQVLSRFLHD
Rv3396c|M.tuberculosis_H37Rv PDGFDVVASSAGAPVAAFEAFDRRLAGVQYHPEVMHTPHGQQVLSRFLHD
MMAR_1148|M.marinum_M PDGFEVVASSAGAAVAAFENRERRLAGVQYHPEVMHTPHGQQILGRFLHD
MUL_0913|M.ulcerans_Agy99 PDGFEVVASSAGAAVAAFENRERRLAGVQYHPEVMHTPHGQQILGRFLHD
MAB_3718c|M.abscessus_ATCC_199 PQGFTVVASSEGAPVAAFENRERRLAGVQYHPEVLHSPHGQQVLSRFLHE
* ** *.*:* *: **.** ************:*:*****:*.****:
Mvan_1516|M.vanbaalenii_PYR-1 FAGIDATWTPANIAEALVEQVRTQIGDGRAICGLSGGVDSAVAAALVQRA
Mflv_4903|M.gilvum_PYR-GCK FAGIDAAWTPANIAEALVEQVRNQIGDGRAICGLSGGVDSAVAAALVQRA
MSMEG_1610|M.smegmatis_MC2_155 FAGIDATWTPANIADQLIEAVREQIGDGRAICGLSGGVDSAVAAALVQRA
TH_0484|M.thermoresistible__bu FAGIGASWTPANIAESLIEQVREQIGDGRAICGLSGGVDSAVAAALVQRA
MLBr_00395|M.leprae_Br4923 FAGLDADWTAANIAGVLVEQVRAQIGNGHAICGLSGGVDSAVAAALVQRA
MAV_4348|M.avium_104 FAGLGADWTAANIAGVLVEQVRAQIGDGHAICGLSGGVDSAVAAALVQRA
Mb3429c|M.bovis_AF2122/97 FAGLGAQWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVAAALVQRA
Rv3396c|M.tuberculosis_H37Rv FAGLGAQWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVAAALVQRA
MMAR_1148|M.marinum_M FAGIGARWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVAAALVQRA
MUL_0913|M.ulcerans_Agy99 FAGIGARWTPANIANALIEQVRTQIGDGHAICGLSGGVDSAVAAALVQRV
MAB_3718c|M.abscessus_ATCC_199 FAGIESAWTAANIAETLVEQVRTQIGEGRALCALSGGVDSAVAAALVQRA
***: : **.**** *:* ** ***:*:*:*.****************.
Mvan_1516|M.vanbaalenii_PYR-1 IGDRLTCVFVDHGLLRSGERAQVQRDFVAATGANLVTVDVADRFLEALSG
Mflv_4903|M.gilvum_PYR-GCK IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDVADRFLEALSG
MSMEG_1610|M.smegmatis_MC2_155 IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGAKLVTVDVADRFLEALTG
TH_0484|M.thermoresistible__bu IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGAKLVTVDTSERFLEALSG
MLBr_00395|M.leprae_Br4923 IGDRLTCIFVDHGLLRDGERGQVQRDFVAATGAKLVTVDAAETFLQALSG
MAV_4348|M.avium_104 IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAADTFLQALAG
Mb3429c|M.bovis_AF2122/97 IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAAETFLEALSG
Rv3396c|M.tuberculosis_H37Rv IGDRLTCVFVDHGLLRAGERAQVQRDFVAATGANLVTVDAAETFLEALSG
MMAR_1148|M.marinum_M IGDRLTCVFVDHGLLRAGERAQVERDFVAATKANLVTVDVADTFLAALSG
MUL_0913|M.ulcerans_Agy99 IGDRLTCVFVDHGLLRAGERAQVERDFVAATKANLVTVDVADTFLAALSG
MAB_3718c|M.abscessus_ATCC_199 IGDRLTCVFVDHGLLRSGERAQVEHDFVAATGATLVTVDASETFLGELAG
*******:******** ***.**::****** *.*****.:: ** *:*
Mvan_1516|M.vanbaalenii_PYR-1 VTNPEGKRKIIGREFIRAFEGAVRDALESQDGS-DVPVEFLVQGTLYPDV
Mflv_4903|M.gilvum_PYR-GCK VTNPEGKRKIIGREFIRAFEGAVRDTVERSDGS-DVPVEFLVQGTLYPDV
MSMEG_1610|M.smegmatis_MC2_155 VTNPEGKRKIIGREFIRAFEGAVRDLVEDEGSG---DIDFLVQGTLYPDV
TH_0484|M.thermoresistible__bu VTNPEGKRKIIGREFIRAFEGAVREALGENAAANDDEVEFLVQGTLYPDV
MLBr_00395|M.leprae_Br4923 VTNPEGKRKIIGRQFIRAFEGAVRDLLGDSTFD--SGIEFLVQGTLYPDV
MAV_4348|M.avium_104 VTNPEGKRKIIGRQFIRAFEGAVRDVLGD------KPVEFLVQGTLYPDV
Mb3429c|M.bovis_AF2122/97 VSAPEGKRKIIGRQFIRAFEGAVRDVLDG------KTAEFLVQGTLYPDV
Rv3396c|M.tuberculosis_H37Rv VSAPEGKRKIIGRQFIRAFEGAVRDVLDG------KTAEFLVQGTLYPDV
MMAR_1148|M.marinum_M VTDPEGKRKIIGRQFIRAFEGAVRDVLDG------RDIEFLVQGTLYPDV
MUL_0913|M.ulcerans_Agy99 VTDPEGKRKIIGRQFIRAFEGAVRDVLDG------RDVEFLVQGTLYPDV
MAB_3718c|M.abscessus_ATCC_199 VTDPEAKRKIIGREFIRSFEGAVSDVVGDSRRDDTAGIQFLVQGTLYPDV
*: **.*******:***:***** : : :***********
Mvan_1516|M.vanbaalenii_PYR-1 VESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRAVGRELGL
Mflv_4903|M.gilvum_PYR-GCK VESGGGSGTANIKSHHNVGGLPDDLKFALVEPLRLLFKDEVRAVGRELGL
MSMEG_1610|M.smegmatis_MC2_155 VESGGGSGTANIKSHHNVGGLPDDLKFKLVEPLRLLFKDEVRAVGRELGL
TH_0484|M.thermoresistible__bu VESGGGAGTANIKSHHNVGGLPDDLKFKLVEPLRLLFKDEVRAVGRELGL
MLBr_00395|M.leprae_Br4923 VESGGGSGTANIKSHHNVGGLPDNLRFKLVEPLRLLFKDEVRAVGRQLDL
MAV_4348|M.avium_104 VESGRGSGTANIKSHHNVGGLPGDLKFTLVEPLRLLFKDEVRAVGRELGL
Mb3429c|M.bovis_AF2122/97 VESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRAVGRELGL
Rv3396c|M.tuberculosis_H37Rv VESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRAVGRELGL
MMAR_1148|M.marinum_M VESGGGSGTANIKSHHNVGGLPDDLKFALVEPLRLLFKDEVRAVGRELGL
MUL_0913|M.ulcerans_Agy99 VESGGGSGTANIKSHHNVGGLPDDLKFTLVEPLRLLFKDEVRAVGRELGL
MAB_3718c|M.abscessus_ATCC_199 VESGGGSGTANIKSHHNVGGLPEDLTFTLVEPLRLLFKDEVRAVGRELGL
**** *:*************** :* * ******************:*.*
Mvan_1516|M.vanbaalenii_PYR-1 PEEIVARQPFPGPGLGIRIVGEVTAERLDTLRRADAIAREELTSAGLDHQ
Mflv_4903|M.gilvum_PYR-GCK PEEIVARQPFPGPGLGIRIIGEVTSDRLDTLRRADVIAREELTAAGLDHQ
MSMEG_1610|M.smegmatis_MC2_155 PEEIVARQPFPGPGLGIRIVGEVTAERLETLRRADAIAREELTAAGLDNQ
TH_0484|M.thermoresistible__bu PEEIVARQPFPGPGLAIRIVGEVTAERLETLRRADAIVREELTAAGLDHQ
MLBr_00395|M.leprae_Br4923 PEEIVARQPFPGPGLGIRIVGEVTAERLDTLRRADSIAREELTTAGLDYQ
MAV_4348|M.avium_104 PEEIVARQPFPGPGLGIRIVGEVTATRLDTLRRADSIAREELTAAGLDSL
Mb3429c|M.bovis_AF2122/97 PEEIVARQPFPGPGLGIRIVGEVTAKRLDTLRHADSIVREELTAAGLDNQ
Rv3396c|M.tuberculosis_H37Rv PEEIVARQPFPGPGLGIRIVGEVTAKRLDTLRHADSIVREELTAAGLDNQ
MMAR_1148|M.marinum_M PEEIVARQPFPGPGLGIRIVGEVTGQRLDTLRRADSIAREELTAAGLDNQ
MUL_0913|M.ulcerans_Agy99 PEEIVARQPFPGPGLGIRIVGEVTGQRLDTLRRADSIARKELTAAGLDNQ
MAB_3718c|M.abscessus_ATCC_199 PEEIVGRQPFPGPGLAIRIVGEVTPDRLDTLRRADAIAREELTAAGQDRN
*****.*********.***:**** **:***:** *.*:***:** *
Mvan_1516|M.vanbaalenii_PYR-1 IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEV
Mflv_4903|M.gilvum_PYR-GCK IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEV
MSMEG_1610|M.smegmatis_MC2_155 IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEV
TH_0484|M.thermoresistible__bu IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYDV
MLBr_00395|M.leprae_Br4923 IWQCPVVLLADVRSVGVQGDNRSYGHPIVLRPVSSEDAMTADWTWVPYEV
MAV_4348|M.avium_104 IWQCPVVLLGEVRSVGVQGDNRTYGHPIVLRPVTSEDAMTADWTRVPYEV
Mb3429c|M.bovis_AF2122/97 IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEV
Rv3396c|M.tuberculosis_H37Rv IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRVPYEV
MMAR_1148|M.marinum_M IWQCPVVLLADVRSVGVQGDNRTYGHPIVLRPVSSEDAMTADWTRVPFEV
MUL_0913|M.ulcerans_Agy99 IWQCPVVLLADVRSVGVQGDNRTYGHPIVLRPVSSEDAMTADWTRVPFEV
MAB_3718c|M.abscessus_ATCC_199 IWQCPVVLLADVRSVGVQGDGRTYGHPIVLRPVSSEDAMTADWTRLPYEV
*********.:*********.*:**********:********** :*::*
Mvan_1516|M.vanbaalenii_PYR-1 LERISTRITNEVREVNRVVLDVTSKPPGTIEWE
Mflv_4903|M.gilvum_PYR-GCK LERISTRITNEVPEVNRVVLDITSKPPGTIEWE
MSMEG_1610|M.smegmatis_MC2_155 LERISTRITNEVREVNRVVLDITSKPPGTIEWE
TH_0484|M.thermoresistible__bu LERISTRITNEVAEVNRVVLDITSKPPATIEWE
MLBr_00395|M.leprae_Br4923 LECISTRITNEVAEVNRVVLDITNKPPGTIEWE
MAV_4348|M.avium_104 LERISTRITNEVPEVNRVVLDITSKPPGTIEWE
Mb3429c|M.bovis_AF2122/97 LERISTRITNEVAEVNRVVLDITSKPPATIEWE
Rv3396c|M.tuberculosis_H37Rv LERISTRITNEVAEVNRVVLDITSKPPATIEWE
MMAR_1148|M.marinum_M LERISTRITNEVPEVNRVVLDVTSKPPGTIEWE
MUL_0913|M.ulcerans_Agy99 LERISTRITNEVPEVNRVVLDVTSKPPGTIEWE
MAB_3718c|M.abscessus_ATCC_199 LERISTRITNEVPEVNRVVLDITSKPPGTIEWE
** ********* ********:*.***.*****