For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RPFDRQKDPCARIRRPPLHTLTGMADSDRLYFRQLLSGRDYAAGDMIAQQMRNFAYLIGDRETGDTVVVD PAYAAQDLVDALEADGMQLSGVLVTHHHPDHIGGSMMGFELKGLAELLERVSVPIHVNAHEADWVSRTTG IARSELTAHQHGDKVSVGDIDIELLHTPGHTPGSQCFLLDGRLVAGDTLFLEGCGRTDFPGGNVDDMFRS LQALAALPGDPTVFPGHWYSADPSAPLSDVRQTNYVYRASDLDQWRMLMGG*
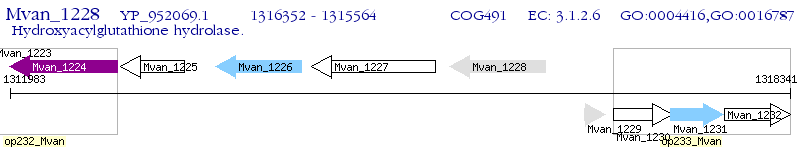
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1228 | - | - | 100% (262) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2589 | - | 7e-17 | 31.22% (189) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_3340 | - | 2e-13 | 28.49% (186) | beta-lactamase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0651c | - | 1e-116 | 82.48% (234) | glyoxalase II |
| M. gilvum PYR-GCK | Mflv_5123 | - | 1e-129 | 91.18% (238) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv0634c | - | 1e-116 | 82.48% (234) | glyoxalase II |
| M. leprae Br4923 | MLBr_01912 | - | 1e-113 | 80.08% (236) | putative glyoxylase II |
| M. abscessus ATCC 19977 | MAB_4755c | - | 4e-15 | 30.94% (181) | hypothetical protein MAB_4755c |
| M. marinum M | MMAR_0965 | gloB | 1e-119 | 84.03% (238) | glyoxalase II, GloB |
| M. avium 104 | MAV_4532 | - | 1e-117 | 83.33% (234) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_1334 | - | 1e-119 | 83.12% (237) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_1873 | - | 1e-121 | 84.39% (237) | POSSIBLE GLYOXALASE II (HYDROXYACYLGLUTATHIONE HYDROLASE) (GLX |
| M. ulcerans Agy99 | MUL_0715 | gloB | 1e-118 | 83.61% (238) | glyoxalase II, GloB |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1228|M.vanbaalenii_PYR-1 MRPFDRQKDPCARIRRPPLHTLTGMADSDRLYFRQLLSGRDYAAGDMIAQ
Mflv_5123|M.gilvum_PYR-GCK ------------------------MADTDRLYFRQLLSGRDFAAGDMIAQ
MSMEG_1334|M.smegmatis_MC2_155 -----------------MAERTSDPAANDRFYFRQLLSGRDFAGADPIAQ
TH_1873|M.thermoresistible__bu ---------------------VSD---SDRLYFRQLLSGRDFAAGDMMAQ
MMAR_0965|M.marinum_M ------------------------MSDSDRLYFRQLLAGRDFAVNDMIAA
MUL_0715|M.ulcerans_Agy99 ------------------------MSDSDRLYFRQLLSGRDFAVNDMIAA
MLBr_01912|M.leprae_Br4923 ------------------------MSDSDRLYFRQLLSGRDFAVDDVVAT
Mb0651c|M.bovis_AF2122/97 ------------------------MSK-DRLYFRQLLSGRDFAVGDMFAT
Rv0634c|M.tuberculosis_H37Rv ------------------------MSK-DRLYFRQLLSGRDFAVGDMFAT
MAV_4532|M.avium_104 ------------------------MSD-DRLYFRQLLSGRDFAAGDMFAT
MAB_4755c|M.abscessus_ATCC_199 ------------------------------MSGIERVITRGTFELDGGSW
: : : *. * :
Mvan_1228|M.vanbaalenii_PYR-1 QMRNFAYLIGDRETGDTVVVDPAYAAQDLVDALEADGMQLSGVLVTHHHP
Mflv_5123|M.gilvum_PYR-GCK QMRNFAYLIGDRETGDAVVVDPAYAAGELVDALEADGMHLSGVLVTHHHP
MSMEG_1334|M.smegmatis_MC2_155 QMRNFAYFIGDRETGEAVVVDPAYAADELAAALEADGMRLAGVLVTHHHP
TH_1873|M.thermoresistible__bu QMRNFAYLIGDRETGECVVVDPAYAANDLVDAAEADGMRLTGVLVSHHHP
MMAR_0965|M.marinum_M QMRNFAYLIGDRETGDCVVVDPAYAAGDLVNALEADDMHLSGVLVTHHHP
MUL_0715|M.ulcerans_Agy99 QMRNFAYLIGDRETGDCVVVDPAYAAGDLVNALEAGDMHLSGVLVTHHHP
MLBr_01912|M.leprae_Br4923 QMRNFAYLIGDRQTGDCVVVDPAYAAGDLVDALEADGMRLSGVLVTHHHP
Mb0651c|M.bovis_AF2122/97 QMRNFAYLIGDRTTGDCVVVDPAYAAGDLLDALESDDMQLSGVLVTHHHP
Rv0634c|M.tuberculosis_H37Rv QMRNFAYLIGDRTTGDCVVVDPAYAAGDLLDALESDDMQLSGVLVTHHHP
MAV_4532|M.avium_104 QMRNFAYLIGDRQTGDCVVVDPAYAASDLLDTLEADGMHLSGVLVTHHHP
MAB_4755c|M.abscessus_ATCC_199 EVDNNIWIVG--EGDDVVVFDAAHSAQPIIDAVG--GRNVVAVVCTHGHN
:: * :::* .: **.*.*::* : : . .: .*: :* *
Mvan_1228|M.vanbaalenii_PYR-1 DHIGGSMMGFELKGLAELLERVSVPIHVNAHEADWVSRTTGIARSELTAH
Mflv_5123|M.gilvum_PYR-GCK DHVGGSMMGFTLKGLAELLERVSVPVHVNAHEADWVSKVTGIAPSELTTH
MSMEG_1334|M.smegmatis_MC2_155 DHVGGSMMGFTLKGVAELLEHRSVPVHVNSHEADWVSQVTGIARSELSAH
TH_1873|M.thermoresistible__bu DHVGGSMMGFTLKGVAELLERAEVPVHVNTHEADWVSRVTGIPLSDLTAH
MMAR_0965|M.marinum_M DHVGGSMMGFELKGLAELLERVSVPVHVNTHEAVWVSRVTGIPMSDLTTH
MUL_0715|M.ulcerans_Agy99 DHVGGSMMGFELKGLAELLERVSVPVHVNTHEAVWVSRVTGIPMSDLTTH
MLBr_01912|M.leprae_Br4923 DHVGGVMMGFQLKGLTELLERVTVPVHVNTHEALWISRVTGIDIGHLTTH
Mb0651c|M.bovis_AF2122/97 DHVGGSMMGFQLPGLAELLERASVPVHVNTHEALWVSRVTGIPVGDLITH
Rv0634c|M.tuberculosis_H37Rv DHVGGSMMGFQLPGLAELLERASVPVHVNTHEALWVSRVTGIPVGDLITH
MAV_4532|M.avium_104 DHVGGSMMGFTLAGLAELVERAPVPVHVNSHEALWVSRVTGISASDLTAH
MAB_4755c|M.abscessus_ATCC_199 DHIT---------VAPELSQALDAPVLL--HPADDVLWRMTHPDKDFRAV
**: .** : .*: : * * : .: :
Mvan_1228|M.vanbaalenii_PYR-1 QHGDKVSVGDIDIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
Mflv_5123|M.gilvum_PYR-GCK QHGDTVSIGDIDIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
MSMEG_1334|M.smegmatis_MC2_155 EHGDTISVGAIDIELLHTPGHTPGSQCFLVDGR--LVAGDTLFLEGCGRT
TH_1873|M.thermoresistible__bu DHGDKVTVGAVEIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
MMAR_0965|M.marinum_M EHRDKVSVGDIEIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
MUL_0715|M.ulcerans_Agy99 EHRDKVSVGDIEIELLHTPGHTPGSQCFLLDGQ--LVAGDTLFLEGCGRT
MLBr_01912|M.leprae_Br4923 EHRDKLSVGDIEIELLHTPGHTPGSQCFLLDGL--LVAGDTLFLEGCGRT
Mb0651c|M.bovis_AF2122/97 EHGDKVSVGDIDIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
Rv0634c|M.tuberculosis_H37Rv EHGDKVSVGDIDIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
MAV_4532|M.avium_104 ENHDKVSIGDIEIELLHTPGHTPGSQCFLLDGR--LVAGDTLFLEGCGRT
MAB_4755c|M.abscessus_ATCC_199 EDDMVLPVGGLELHALHTPGHSPGSVCWYAPELRAVFSGDTLFQGGPGAT
:. :.:* :::. ******:*** *: :.:***** * * *
Mvan_1228|M.vanbaalenii_PYR-1 DFPGGNVDDMFRSLQ-ALAALPGDPTVFPGHWYSADPSAPLSDVRQTNYV
Mflv_5123|M.gilvum_PYR-GCK DFPGGDVDDMFRSLQ-ALAQLPGDPTVFPGHWYSAEPSAPLSDVRGSNYV
MSMEG_1334|M.smegmatis_MC2_155 DFPGGNVDDMFRSLQ-ALAKLPGDPTVYPGHWYSAEPSAPLEEVKRSNYV
TH_1873|M.thermoresistible__bu DFPGGNVDDMFHSLQ-ALAKLPGDPTVYPGHWYAAEPSASLSEVRRTNYV
MMAR_0965|M.marinum_M DFPGGDSDEIFRSLQ-QLAALPGDPTVFPGHWYSAEPSASLSEVKRSNFV
MUL_0715|M.ulcerans_Agy99 DFPGGDSDEIFRSLQ-QLAALPGDPTVFPGHWYSAEPSASLSEVKRSNFV
MLBr_01912|M.leprae_Br4923 DFPGGNSDEMYRSLQ-QLAQLPGNPTVFPGHWYSTAPSATLSMVKCSNYV
Mb0651c|M.bovis_AF2122/97 DFPGGDSDEMYRSLR-QLAELPGDPTVFPGHWYSAEPSASLSEVKRSNYV
Rv0634c|M.tuberculosis_H37Rv DFPGGDSDEMYRSLR-QLAELPGDPTVFPGHWYSAEPSASLSEVKRSNYV
MAV_4532|M.avium_104 DFPGGDSDEMYRSLQ-RLAKLPGDPTVFPGHWYSAEPSASLSEVKRSNYV
MAB_4755c|M.abscessus_ATCC_199 GRSFSDFPTILRSISGRLGPLPEDTVVYTGHGDTTTIGD--EIVHYDDWV
. . .: : :*: *. ** :..*:.** :: . . *: ::*
Mvan_1228|M.vanbaalenii_PYR-1 YRASDLDQWRMLMGG
Mflv_5123|M.gilvum_PYR-GCK YRASNLDQWRMLMGG
MSMEG_1334|M.smegmatis_MC2_155 YRASNLDQWRMLMGG
TH_1873|M.thermoresistible__bu YRASNLEQWRMLMGA
MMAR_0965|M.marinum_M YRAADLAQWRMLMGG
MUL_0715|M.ulcerans_Agy99 YRAADLAQWRMLMGG
MLBr_01912|M.leprae_Br4923 YRAADLNQWRMLMDS
Mb0651c|M.bovis_AF2122/97 YRPASLDQWRMLMGG
Rv0634c|M.tuberculosis_H37Rv YRPASLDQWRMLMGG
MAV_4532|M.avium_104 YRASDLDQWRMLMGG
MAB_4755c|M.abscessus_ATCC_199 ARGH-----------
*