For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSKDRLYFRQLLSGRDFAVGDMFATQMRNFAYLIGDRTTGDCVVVDPAYAAGDLLDALESDDMQLSGVLV THHHPDHVGGSMMGFQLPGLAELLERASVPVHVNTHEALWVSRVTGIPVGDLITHEHGDKVSVGDIDIEL LHTPGHTPGSQCFLLDGRLVAGDTLFLEGCGRTDFPGGDSDEMYRSLRQLAELPGDPTVFPGHWYSAEPS ASLSEVKRSNYVYRPASLDQWRMLMGG
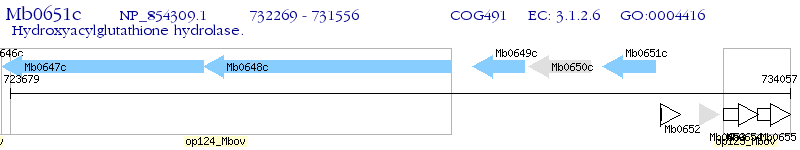
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0651c | - | - | 100% (237) | glyoxalase II |
| M. bovis AF2122 / 97 | Mb2284 | - | 1e-15 | 34.41% (186) | hypothetical protein Mb2284 |
| M. bovis AF2122 / 97 | Mb2612c | - | 3e-15 | 28.93% (197) | glyoxalase II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5123 | - | 1e-117 | 84.19% (234) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv0634c | - | 1e-140 | 100.00% (237) | glyoxalase II |
| M. leprae Br4923 | MLBr_01912 | - | 1e-118 | 85.34% (232) | putative glyoxylase II |
| M. abscessus ATCC 19977 | MAB_2873c | - | 9e-16 | 28.12% (224) | hypothetical protein MAB_2873c |
| M. marinum M | MMAR_0965 | gloB | 1e-124 | 88.89% (234) | glyoxalase II, GloB |
| M. avium 104 | MAV_4532 | - | 1e-124 | 88.19% (237) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_1334 | - | 1e-109 | 78.39% (236) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_1873 | - | 1e-115 | 81.55% (233) | POSSIBLE GLYOXALASE II (HYDROXYACYLGLUTATHIONE HYDROLASE) (GLX |
| M. ulcerans Agy99 | MUL_0715 | gloB | 1e-123 | 88.46% (234) | glyoxalase II, GloB |
| M. vanbaalenii PYR-1 | Mvan_1228 | - | 1e-115 | 82.48% (234) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5123|M.gilvum_PYR-GCK ------------------------MADTDRLYFRQLLSGRDFAAGDMIAQ
Mvan_1228|M.vanbaalenii_PYR-1 MRPFDRQKDPCARIRRPPLHTLTGMADSDRLYFRQLLSGRDYAAGDMIAQ
MSMEG_1334|M.smegmatis_MC2_155 -----------------MAERTSDPAANDRFYFRQLLSGRDFAGADPIAQ
TH_1873|M.thermoresistible__bu ---------------------VSD---SDRLYFRQLLSGRDFAAGDMMAQ
Mb0651c|M.bovis_AF2122/97 ------------------------MSK-DRLYFRQLLSGRDFAVGDMFAT
Rv0634c|M.tuberculosis_H37Rv ------------------------MSK-DRLYFRQLLSGRDFAVGDMFAT
MAV_4532|M.avium_104 ------------------------MSD-DRLYFRQLLSGRDFAAGDMFAT
MMAR_0965|M.marinum_M ------------------------MSDSDRLYFRQLLAGRDFAVNDMIAA
MUL_0715|M.ulcerans_Agy99 ------------------------MSDSDRLYFRQLLSGRDFAVNDMIAA
MLBr_01912|M.leprae_Br4923 ------------------------MSDSDRLYFRQLLSGRDFAVDDVVAT
MAB_2873c|M.abscessus_ATCC_199 ----------------------------------MLITG--FPAG-----
*::* :.
Mflv_5123|M.gilvum_PYR-GCK QMRNFAYLIGDRETGDAVVVDP-AYAAGELVDALEADGMHLSGVLVTHHH
Mvan_1228|M.vanbaalenii_PYR-1 QMRNFAYLIGDRETGDTVVVDP-AYAAQDLVDALEADGMQLSGVLVTHHH
MSMEG_1334|M.smegmatis_MC2_155 QMRNFAYFIGDRETGEAVVVDP-AYAADELAAALEADGMRLAGVLVTHHH
TH_1873|M.thermoresistible__bu QMRNFAYLIGDRETGECVVVDP-AYAANDLVDAAEADGMRLTGVLVSHHH
Mb0651c|M.bovis_AF2122/97 QMRNFAYLIGDRTTGDCVVVDP-AYAAGDLLDALESDDMQLSGVLVTHHH
Rv0634c|M.tuberculosis_H37Rv QMRNFAYLIGDRTTGDCVVVDP-AYAAGDLLDALESDDMQLSGVLVTHHH
MAV_4532|M.avium_104 QMRNFAYLIGDRQTGDCVVVDP-AYAASDLLDTLEADGMHLSGVLVTHHH
MMAR_0965|M.marinum_M QMRNFAYLIGDRETGDCVVVDP-AYAAGDLVNALEADDMHLSGVLVTHHH
MUL_0715|M.ulcerans_Agy99 QMRNFAYLIGDRETGDCVVVDP-AYAAGDLVNALEAGDMHLSGVLVTHHH
MLBr_01912|M.leprae_Br4923 QMRNFAYLIGDRQTGDCVVVDP-AYAAGDLVDALEADGMRLSGVLVTHHH
MAB_2873c|M.abscessus_ATCC_199 MFATNCYIVAPHEGGPAVIVDPGQDAIGGVKEILAERDLTPEAVLLTHGH
: . .*::. : * *:*** * : .: .**::* *
Mflv_5123|M.gilvum_PYR-GCK PDHVGGSMMGFTLKGLAELLER------VSVPVHVNAHEADWVSKVTGIA
Mvan_1228|M.vanbaalenii_PYR-1 PDHIGGSMMGFELKGLAELLER------VSVPIHVNAHEADWVSRTTGIA
MSMEG_1334|M.smegmatis_MC2_155 PDHVGGSMMGFTLKGVAELLEH------RSVPVHVNSHEADWVSQVTGIA
TH_1873|M.thermoresistible__bu PDHVGGSMMGFTLKGVAELLER------AEVPVHVNTHEADWVSRVTGIP
Mb0651c|M.bovis_AF2122/97 PDHVGGSMMGFQLPGLAELLER------ASVPVHVNTHEALWVSRVTGIP
Rv0634c|M.tuberculosis_H37Rv PDHVGGSMMGFQLPGLAELLER------ASVPVHVNTHEALWVSRVTGIP
MAV_4532|M.avium_104 PDHVGGSMMGFTLAGLAELVER------APVPVHVNSHEALWVSRVTGIS
MMAR_0965|M.marinum_M PDHVGGSMMGFELKGLAELLER------VSVPVHVNTHEAVWVSRVTGIP
MUL_0715|M.ulcerans_Agy99 PDHVGGSMMGFELKGLAELLER------VSVPVHVNTHEAVWVSRVTGIP
MLBr_01912|M.leprae_Br4923 PDHVGGVMMGFQLKGLTELLER------VTVPVHVNTHEALWISRVTGID
MAB_2873c|M.abscessus_ATCC_199 LDHTWTAQPLADEYGIPVYIHPDDRVMLADPLAGIGPGLGQFIQGQVFAE
** *:. :. :.. . ::. .
Mflv_5123|M.gilvum_PYR-GCK PSELTTHQHGDTVSIGDIDIELLHTPGHTPGSQCFLLDG--------RLV
Mvan_1228|M.vanbaalenii_PYR-1 RSELTAHQHGDKVSVGDIDIELLHTPGHTPGSQCFLLDG--------RLV
MSMEG_1334|M.smegmatis_MC2_155 RSELSAHEHGDTISVGAIDIELLHTPGHTPGSQCFLVDG--------RLV
TH_1873|M.thermoresistible__bu LSDLTAHDHGDKVTVGAVEIELLHTPGHTPGSQCFLLDG--------RLV
Mb0651c|M.bovis_AF2122/97 VGDLITHEHGDKVSVGDIDIELLHTPGHTPGSQCFLLDG--------RLV
Rv0634c|M.tuberculosis_H37Rv VGDLITHEHGDKVSVGDIDIELLHTPGHTPGSQCFLLDG--------RLV
MAV_4532|M.avium_104 ASDLTAHENHDKVSIGDIEIELLHTPGHTPGSQCFLLDG--------RLV
MMAR_0965|M.marinum_M MSDLTTHEHRDKVSVGDIEIELLHTPGHTPGSQCFLLDG--------RLV
MUL_0715|M.ulcerans_Agy99 MSDLTTHEHRDKVSVGDIEIELLHTPGHTPGSQCFLLDG--------QLV
MLBr_01912|M.leprae_Br4923 IGHLTTHEHRDKLSVGDIEIELLHTPGHTPGSQCFLLDG--------LLV
MAB_2873c|M.abscessus_ATCC_199 PKELVEIGDGDKLELAGITLTVDHTPGHTRGSVVFGIEVDSENGPVDVLF
.* . *.: :. : : : ****** ** * :: *.
Mflv_5123|M.gilvum_PYR-GCK AGDTLFLEGCGRTDFPGGDVDDMFRSLQALAQLPGDP-TVFPGHWYSAEP
Mvan_1228|M.vanbaalenii_PYR-1 AGDTLFLEGCGRTDFPGGNVDDMFRSLQALAALPGDP-TVFPGHWYSADP
MSMEG_1334|M.smegmatis_MC2_155 AGDTLFLEGCGRTDFPGGNVDDMFRSLQALAKLPGDP-TVYPGHWYSAEP
TH_1873|M.thermoresistible__bu AGDTLFLEGCGRTDFPGGNVDDMFHSLQALAKLPGDP-TVYPGHWYAAEP
Mb0651c|M.bovis_AF2122/97 AGDTLFLEGCGRTDFPGGDSDEMYRSLRQLAELPGDP-TVFPGHWYSAEP
Rv0634c|M.tuberculosis_H37Rv AGDTLFLEGCGRTDFPGGDSDEMYRSLRQLAELPGDP-TVFPGHWYSAEP
MAV_4532|M.avium_104 AGDTLFLEGCGRTDFPGGDSDEMYRSLQRLAKLPGDP-TVFPGHWYSAEP
MMAR_0965|M.marinum_M AGDTLFLEGCGRTDFPGGDSDEIFRSLQQLAALPGDP-TVFPGHWYSAEP
MUL_0715|M.ulcerans_Agy99 AGDTLFLEGCGRTDFPGGDSDEIFRSLQQLAALPGDP-TVFPGHWYSAEP
MLBr_01912|M.leprae_Br4923 AGDTLFLEGCGRTDFPGGNSDEMYRSLQQLAQLPGNP-TVFPGHWYSTAP
MAB_2873c|M.abscessus_ATCC_199 SGDTLFQMSIGRTDLPGGNHQQLLDSIAAKLLTRDDSSVVLPGHGN----
:***** . ****:***: ::: *: .:. .* ***
Mflv_5123|M.gilvum_PYR-GCK SAPLSDVRGSNYVYRASNLDQWRMLMGG
Mvan_1228|M.vanbaalenii_PYR-1 SAPLSDVRQTNYVYRASDLDQWRMLMGG
MSMEG_1334|M.smegmatis_MC2_155 SAPLEEVKRSNYVYRASNLDQWRMLMGG
TH_1873|M.thermoresistible__bu SASLSEVRRTNYVYRASNLEQWRMLMGA
Mb0651c|M.bovis_AF2122/97 SASLSEVKRSNYVYRPASLDQWRMLMGG
Rv0634c|M.tuberculosis_H37Rv SASLSEVKRSNYVYRPASLDQWRMLMGG
MAV_4532|M.avium_104 SASLSEVKRSNYVYRASDLDQWRMLMGG
MMAR_0965|M.marinum_M SASLSEVKRSNFVYRAADLAQWRMLMGG
MUL_0715|M.ulcerans_Agy99 SASLSEVKRSNFVYRAADLAQWRMLMGG
MLBr_01912|M.leprae_Br4923 SATLSMVKCSNYVYRAADLNQWRMLMDS
MAB_2873c|M.abscessus_ATCC_199 ATSIGDERRANPFLEGIQ----------
::.: : :* . . .