For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQCVGALRLDGALALSDHGRTASSDDRARREALAEFLRARRESLQPEDIGIPRRGRRRVKGLRRHEVADA AAVSVTWYTWLEQGRDIHATPQVIEALARALQLDDSEHSYLCRLAGVTPKATQLGRTQAGDSLIALVDSL LPNPAQLMLPATDLINWNDAYSRLFVDPSTLAAEDRNALWIQVMCPEVRHRLVDWEAETQLVVGRFRAEA AKYPGDPHFRRIIDRLSAESTLFRSIWARHEVQGFEDRVKTIEHPEVGEVRVRLVKLRPLDHPHLLLLVH MLDDDESRARIGRLLSR
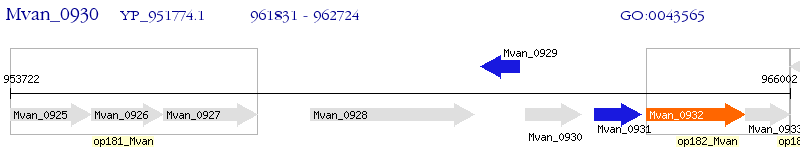
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0930 | - | - | 100% (297) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1162 | - | 6e-49 | 37.81% (283) | XRE family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5465 | - | 6e-33 | 36.97% (238) | XRE family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5170 | - | 6e-48 | 36.63% (273) | XRE family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4641 | - | 1e-50 | 40.59% (271) | putative DNA-binding protein |
| M. marinum M | MMAR_3947 | - | 1e-05 | 28.40% (162) | transcriptional regulatory protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1289 | - | 9e-51 | 41.50% (253) | hypothetical protein MSMEG_1289 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4641|M.abscessus_ATCC_1997 --------------------MAGTDE--RRRELGAFLRVRREHLVRADYG
MSMEG_1289|M.smegmatis_MC2_155 --------------------MATQEE--HRAELGAFLRASRERLNRAEFG
Mflv_5170|M.gilvum_PYR-GCK --------------------MDTRAQ--RRLALGEFLRARREAIRRADVG
Mvan_0930|M.vanbaalenii_PYR-1 MQCVGALRLDGALALSDHGRTASSDDRARREALAEFLRARRESLQPEDIG
MMAR_3947|M.marinum_M -----------------------------MPNRGGFVHAARER-------
. *::. **
MAB_4641|M.abscessus_ATCC_1997 LPPVGRSRTTGLRREEIAYLSGVSVTWYTWLEQGRDINPSRQVLEAIALE
MSMEG_1289|M.smegmatis_MC2_155 LPPAGRNRTVGLRREEVSYLSGVSVTWYTWLEQGRDINPSRQVLDAVSRT
Mflv_5170|M.gilvum_PYR-GCK LPELPRARTGGLRREEVAVTAGVSVTWYTWLEQGRDINPSKQVLDAVADA
Mvan_0930|M.vanbaalenii_PYR-1 IPRRGRRRVKGLRRHEVADAAAVSVTWYTWLEQGRDIHATPQVIEALARA
MMAR_3947|M.marinum_M ---------AGLTCRELAEKAGCSLRYVHDIEQG--CSPSIRVLNGLVRA
** .*:: :. *: : :*** .: :*::.:
MAB_4641|M.abscessus_ATCC_1997 MRLSAAERTYLLELAGFAPLPHSATPDPAPVPDHLLRLVENLVPSPAFVV
MSMEG_1289|M.smegmatis_MC2_155 LRLSAAQHRYVLTLAGFAPVSAAPADDVTDVPGHVQRLLDAIGDNPAYAL
Mflv_5170|M.gilvum_PYR-GCK LRLSPAEHEYVLALGGYA---VDAAVSRGPLPPHIQHFLDALEGYPAFAI
Mvan_0930|M.vanbaalenii_PYR-1 LQLDDSEHSYLCRLAGVTP--KATQLGRTQAGDSLIALVDSLLPNPAQLM
MMAR_3947|M.marinum_M LQLSAWEQRFLFAACGRILP--DPALTDTDIS---QRYLDALQPHPAAAL
::*. :: :: * . :: : ** :
MAB_4641|M.abscessus_ATCC_1997 SADWSIGGWNRAYELLYPGIASIVPADRNLLRFIFTDPYVRQMLPDWPIT
MSMEG_1289|M.smegmatis_MC2_155 TPEWGIAGWNAAYQRLYPNVATTPPAERNLLWAVFTDPFVRDLLDDWPVT
Mflv_5170|M.gilvum_PYR-GCK TPDWGIAAWNTTYAALYPNVERVPETDRNLLWLVFTDPYIHELLPNWDTD
Mvan_0930|M.vanbaalenii_PYR-1 LPATDLINWNDAYSRLFVDPSTLAAEDRNALWIQVMCPEVRHRLVDWEAE
MMAR_3947|M.marinum_M HPSWRFKAWNQQWQRLFQGVLLAPS----MHQWLYLSTTSRRILRNWDEV
. : ** : *: . . : * :*
MAB_4641|M.abscessus_ATCC_1997 SRRFLAEYRAEAGALLGDPRHRALVDRLQQQSPEFARAWAEHEVGRFTSR
MSMEG_1289|M.smegmatis_MC2_155 SQRFLAEFRAEIGPRLGNPTLLGLIERLSEASDEFRAGWERHDIRGFESR
Mflv_5170|M.gilvum_PYR-GCK SRRFLAEFRADAGPRVGEPSYTALVSRLSSESPHFAAAWQATSIERFTSR
Mvan_0930|M.vanbaalenii_PYR-1 TQLVVGRFRAEAAKYPGDPHFRRIIDRLSAESTLFRSIWARHEVQGFEDR
MMAR_3947|M.marinum_M SQWCVGWLRYGLAVDVD--AVKPMVSALMP-VTEFRREWEAQVIPVDPAT
:: :. * . . ::. * * * :
MAB_4641|M.abscessus_ATCC_1997 ERRFHHPEAGELIFEHHRLSPSDLPDLHIVIYLPLDGTNTRTKLEALLSG
MSMEG_1289|M.smegmatis_MC2_155 ERVFHHPEFGVVTYEHHQLRPSDQPELQLVVYTPTHRNP-----------
Mflv_5170|M.gilvum_PYR-GCK ERLFRHPAAGELRLEHHQVAPVDVPDIQIVAYLPVPDSVASERLRMLTGQ
Mvan_0930|M.vanbaalenii_PYR-1 VKTIEHPEVGEVRVRLVKLRPLDHPHLLLLVHM-LDDDESRARIGRLLSR
MMAR_3947|M.marinum_M RLWVVRDGGTELRIDMRFWHASPMPGGLLLGVILDAG-------------
. : : . * ::
MAB_4641|M.abscessus_ATCC_1997 GR---
MSMEG_1289|M.smegmatis_MC2_155 -----
Mflv_5170|M.gilvum_PYR-GCK ADTAR
Mvan_0930|M.vanbaalenii_PYR-1 -----
MMAR_3947|M.marinum_M -----